Fibroblast hierarchy dynamics during mammary gland morphogenesis and tumorigenesis
- PMID: 40216939
- PMCID: PMC12130467
- DOI: 10.1038/s44318-025-00422-3
Fibroblast hierarchy dynamics during mammary gland morphogenesis and tumorigenesis
Abstract
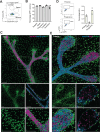
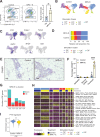
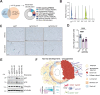
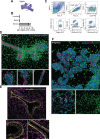
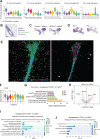
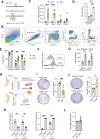
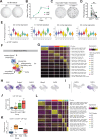
Fibroblasts form a major component of the stroma in normal mammary tissue and breast tumors. Here, we have applied longitudinal single-cell transcriptome profiling of >45,000 fibroblasts in the mouse mammary gland across five different developmental stages and during oncogenesis. In the normal gland, diverse stromal populations were resolved, including lobular-like fibroblasts, committed preadipocytes and adipogenesis-regulatory, as well as cycling fibroblasts in puberty and pregnancy. These specialized cell types appear to emerge from CD34high mesenchymal progenitor cells, accompanied by elevated Hedgehog signaling. During late tumorigenesis, heterogeneous cancer-associated fibroblasts (CAFs) were identified in mouse models of breast cancer, including a population of CD34- myofibroblastic CAFs (myCAFs) that were transcriptionally and phenotypically similar to senescent CAFs. Moreover, Wnt9a was demonstrated to be a regulator of senescence in CD34- myCAFs. These findings reflect a diverse and hierarchically organized stromal compartment in the normal mammary gland that provides a framework to better understand fibroblasts in normal and cancerous states.
Keywords: Cancer-Associated Fibroblasts (CAFs); Fibroblasts; Mammary Gland Development; Senescence; Wnt9a.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures






References
-
- Abbasi S, Sinha S, Labit E, Rosin NL, Yoon G, Rahmani W, Jaffer A, Sharma N, Hagner A, Shah P et al (2020) Distinct regulatory programs control the latent regenerative potential of dermal fibroblasts during wound healing. Cell Stem Cell 27:396–412.e396 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

