The role of hypoxia-senescence co-related molecular subtypes and prognostic characteristics in hepatocellular carcinoma
- PMID: 40216977
- PMCID: PMC11992139
- DOI: 10.1038/s41598-025-97604-0
The role of hypoxia-senescence co-related molecular subtypes and prognostic characteristics in hepatocellular carcinoma
Abstract
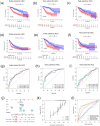
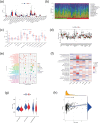
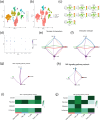
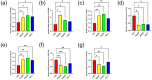
Hepatocellular carcinoma (HCC) is known for its high invasiveness, high fatality rate. Both hypoxia and senescence play crucial roles in the initiation and progression of cancer, yet their prognostic implications in HCC are yet to be fully understood. The hypoxia-senescence co-related genes (HSCRGs) were screened from public databases. Transcriptome data and clinical information were obtained from patients with HCC using the Cancer Genome Atlas, GSE76427, and International Cancer Genome Consortium (ICGC). The random forest tree algorithm was used to identify the characteristic genes of the disease, and the genes were verified by related experiments. SVM algorithm was used to classify HCC patients based on HSCRGs. The prediction model based on HSCRGs was established by LASSO, univariate and multivariate COX regression analysis. We used the ICGC for outside validation. The risk score model was analyzed from subgroup analysis, immune infiltration, and functional strength. The expression patterns of key prognostic genes in tumor microenvironment were decoded by single cell analysis. A total of 184 HSCRGs were identified. The expression pattern and functional characteristics of MLH1 gene in HCC were verified. Two HCC subtypes were identified based on HSCRGs. Then, a prediction model based on HSCRGs was established, and risk score was identified as an independent prognostic indicator of HCC. A new nomogram is constructed and shows good prediction ability. We further determined that the level of infiltration of immune cells and the expression of immune checkpoints are significantly affected by the risk score. The immune microenvironment was different between the two risk groups. The high-risk group was dominated by immunosuppressed cells, and the prognosis was poor. Single-cell analysis revealed the expression of seven key prognostic genes in the tumor microenvironment. Finally, qPCR results further verified the expression levels of seven prognostic genes. HSCRGs are of great significance in the prognosis prediction, risk stratification and targeted therapy of patients with HCC.
Keywords: Hepatocellular carcinoma; Hypoxia; MLH1; Molecular subtypes; Prognostic risk model; Senescence.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
A m6A regulators-related classifier for prognosis and tumor microenvironment characterization in hepatocellular carcinoma.Front Immunol. 2024 Jul 25;15:1374465. doi: 10.3389/fimmu.2024.1374465. eCollection 2024. Front Immunol. 2024. PMID: 39119345 Free PMC article.
-
Prognostic modeling of hepatocellular carcinoma based on T-cell proliferation regulators: a bioinformatics approach.Front Immunol. 2024 Oct 9;15:1444091. doi: 10.3389/fimmu.2024.1444091. eCollection 2024. Front Immunol. 2024. PMID: 39445019 Free PMC article.
-
Construction of a risk score prognosis model based on hepatocellular carcinoma microenvironment.World J Gastroenterol. 2020 Jan 14;26(2):134-153. doi: 10.3748/wjg.v26.i2.134. World J Gastroenterol. 2020. PMID: 31969776 Free PMC article.
-
Identification of drivers from cancer genome diversity in hepatocellular carcinoma.Int J Mol Sci. 2014 Jun 20;15(6):11142-60. doi: 10.3390/ijms150611142. Int J Mol Sci. 2014. PMID: 24955791 Free PMC article. Review.
-
Cytokine biomarkers for independent prediction of hepatocellular carcinoma prognosis.Discov Oncol. 2025 Mar 29;16(1):421. doi: 10.1007/s12672-025-02188-w. Discov Oncol. 2025. PMID: 40155531 Free PMC article. Review.
References
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249. 10.3322/caac.21660 (2021). - PubMed
-
- Ali, N. A., Hamdy, N. M., Gibriel, A. A. & El Mesallamy, H. O. Investigation of the relationship between CTLA4 and the tumor suppressor RASSF1A and the possible mediating role of STAT4 in a cohort of Egyptian patients infected with hepatitis C virus with and without hepatocellular carcinoma. Arch. Virol.166, 1643–1651. 10.1007/s00705-021-04981-8 (2021). - PubMed
-
- Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Primers7, 6. 10.1038/s41572-020-00240-3 (2021). - PubMed
-
- Youssef, S. S. & Hamdy, N. M. SOCS1 and pattern recognition receptors: TLR9 and RIG-I; novel haplotype associations in Egyptian fibrotic/cirrhotic patients with HCV genotype 4. Arch. Virol.162, 3347–3354. 10.1007/s00705-017-3498-7 (2017). - PubMed
-
- Elanany, M. M., Mostafa, D. & Hamdy, N. M. Remodeled tumor immune microenvironment (TIME) parade via natural killer cells reprogramming in breast cancer. Life Sci.330, 121997. 10.1016/j.lfs.2023.121997 (2023). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

