Pre-symptomatic Parkinson's disease blood test quantifying repetitive sequence motifs in transfer RNA fragments
- PMID: 40216989
- PMCID: PMC12092246
- DOI: 10.1038/s43587-025-00851-z
Pre-symptomatic Parkinson's disease blood test quantifying repetitive sequence motifs in transfer RNA fragments
Abstract
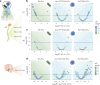
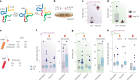
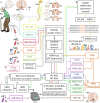
Early, efficient Parkinson's disease (PD) tests may facilitate pre-symptomatic diagnosis and disease-modifying therapies. Here we report elevated levels of PD-specific transfer RNA fragments carrying a conserved sequence motif (RGTTCRA-tRFs) in the substantia nigra, cerebrospinal fluid and blood of patients with PD. A whole blood qPCR test detecting elevated RGTTCRA-tRFs and reduced mitochondrial-originated tRFs (MT-tRFs) segregated pre-symptomatic patients with PD from controls (area under the receiver operating characteristic curve (ROC-AUC) of 0.75 versus 0.71 based on traditional clinical scoring). Strengthening PD relevance, patients carrying PD-related mutations presented higher blood RGTTCRA-tRFs/MT-tRFs ratios than mutation-carrying non-symptomatic controls, and RGTTCRA-tRF levels decreased in patients' blood after deep brain stimulation. Furthermore, RGTTCRA-tRFs complementarity to ribosomal RNA and the translation-supporting LeuCAG3-tRF might aggravate PD via translational inhibition, as reflected by disrupted ribosomal association of RGTTCRA-tRFs in depolarized neuroblastoma cells. Our findings show tRF involvement in PD and suggest a potential simple and safe blood test that may aid clinicians in pre-symptomatic PD diagnosis after validation in larger independent cohorts.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.S., N.M., E.L. and S.V.-T. have patents listed on the finding of the PD motif sequence: application 63/531,628, titled ‘Methods for detection, qualification, or monitoring of Parkinson’s disease’, and application number 63/557,661, titled ‘Angiogenin-reducing agents for treating Parkinson’s disease’. H.S. serves as the senior academic advisor to the Azrieli Fellows Program in Israel. The other authors declare no competing interests.
Figures








References
-
- Aarsland, D. et al. Parkinson disease-associated cognitive impairment. Nat. Rev. Dis. Primers7, 47 (2021). - PubMed
-
- Parkinson’s disease economic burden on patients, families and the federal government is $52 billion, doubling previous estimates. https://www.michaeljfox.org/publication/parkinsons-disease-economic-burd... (2019).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

