Targeting PRDX2 to inhibit tumor growth and metastasis in triple-negative breast cancer: the role of FN1 and the PI3K/AKT/SP1 pathway
- PMID: 40217291
- PMCID: PMC11992733
- DOI: 10.1186/s12967-025-06441-2
Targeting PRDX2 to inhibit tumor growth and metastasis in triple-negative breast cancer: the role of FN1 and the PI3K/AKT/SP1 pathway
Abstract
Background: Triple-negative breast cancer (TNBC) is characterized by high invasiveness, high potential for metastasis, easy recurrence, and poor prognosis. There is an urgent need to develop new clinical treatments.
Methods: This study utilized TNBC tissue microarrays to detect Peroxiredoxin 2 (PRDX2) expression levels and analyzed the correlation between PRDX2 and tumor invasion as well as invasion-related gene expression through the TCGA database. A stable PRDX2-knockdown triple-negative breast cancer cell line was established using lentiviral technology. The effects of PRDX2 on triple-negative breast cancer cell migration, invasion, and epithelial-mesenchymal transition (EMT) were investigated via wound healing assays, Transwell assays, qPCR, and Western blotting. RNA sequencing (RNA-seq), Western blotting, and dual luciferase reporter assays were performed to confirm that PRDX2 regulates FN1 expression through SP1. Furthermore, subcutaneous tumor xenograft models in nude mice were constructed to assess the effects of PRDX2 knockdown and the PRDX2 inhibitor Conoidin A on tumor growth in vivo.
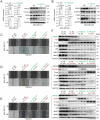
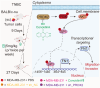
Results: Tissue microarray detection and correlative analysis revealed that PRDX2 is significantly upregulated in triple-negative breast cancer (TNBC) tumor tissues and positively correlated with genes associated with cell migration and invasion. Functional experiments demonstrated that in vitro knockdown of PRDX2 suppresses migration, invasion, and epithelial-mesenchymal transition (EMT) in TNBC cells. Furthermore, in vivo knockdown of PRDX2 or treatment with the PRDX2 inhibitor Conoidin A effectively reduced tumor burden. Mechanistic investigations utilizing RNA sequencing (RNA-seq) identified FN1 as a critical gene promoting TNBC cell migration and invasion. PRDX2 facilitates TNBC progression by activating the PI3K/AKT signaling pathway, which enhances SP1 binding to the FN1 gene promoter. This regulatory cascade ultimately drives tumor advancement in TNBC.
Conclusions: This study elucidates the role of the PRDX2/SP1/FN1 axis in TNBC migration and invasion, and highlights PRDX2 as a promising therapeutic target for triple-negative breast cancer.
Keywords: Cell migration and invasion; PI3K/AKT; PRDX2; SP1; TNBC.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The ethics involving human tissues (tissue microarrays of triple-negative breast cancer (TNBC) patients) were approved by the Ethics Committee of Shanghai New Tide Biotechnology Co., Ltd. (approval No. SHYJS-CP-2210008). The ethics involving animals was approved by the medical ethics committee of Anhui University of Science and Technology (approval NO. HX-002). Consent for publication: All the authors approved the publication. Competing interests: The authors declare that they have no competing interests.
Figures




Similar articles
-
LINC01614 activated by SP1 promoted malignant behavior of triple-negative breast cancer cells via the WNT/b-Catenin signaling pathway.Rev Invest Clin. 2024 Oct 17;76(4):185-194. doi: 10.24875/RIC.24000093. Rev Invest Clin. 2024. PMID: 39419019
-
Long noncoding RNA AFAP1-AS1 promotes tumor progression and invasion by regulating the miR-2110/Sp1 axis in triple-negative breast cancer.Cell Death Dis. 2021 Jun 18;12(7):627. doi: 10.1038/s41419-021-03917-z. Cell Death Dis. 2021. PMID: 34145213 Free PMC article.
-
Calycosin inhibits triple-negative breast cancer progression through down-regulation of the novel estrogen receptor-α splice variant ER-α30-mediated PI3K/AKT signaling pathway.Phytomedicine. 2023 Sep;118:154924. doi: 10.1016/j.phymed.2023.154924. Epub 2023 Jun 14. Phytomedicine. 2023. PMID: 37393829
-
Divide and Conquer-Targeted Therapy for Triple-Negative Breast Cancer.Int J Mol Sci. 2025 Feb 7;26(4):1396. doi: 10.3390/ijms26041396. Int J Mol Sci. 2025. PMID: 40003864 Free PMC article. Review.
-
miRNA signatures affecting the survival outcome in distant metastasis of triple-negative breast cancer.Biochem Pharmacol. 2025 Jan;231:116683. doi: 10.1016/j.bcp.2024.116683. Epub 2024 Nov 27. Biochem Pharmacol. 2025. PMID: 39608504 Review.
References
-
- Harbeck N, Gnant M. Breast cancer Lancet. 2017;389:1134–50. - PubMed
-
- Zou Y, Xie J, Zheng S, Liu W, Tang Y, Tian W, Deng X, et al. Leveraging diverse cell-death patterns to predict the prognosis and drug sensitivity of triple-negative breast cancer patients after surgery. Int J Surg. 2022;107:106936. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

