Decoding mitochondrial stress genes in DCM: towards precision diagnosis and therapy
- PMID: 40217309
- PMCID: PMC11987231
- DOI: 10.1186/s41065-025-00399-3
Decoding mitochondrial stress genes in DCM: towards precision diagnosis and therapy
Abstract
Background: Mitochondrial oxidative stress (ROS) is a crucial factor in the pathogenesis of dilated cardiomyopathy (DCM). Despite its significance, robust biomarkers for assessing its role remain scarce. This study investigates ROS mechanisms in DCM and identifies associated biomarkers, offering fresh insights into diagnosis and treatment.
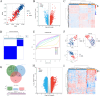
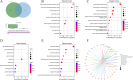
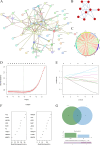
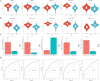
Methods: We sourced transcriptomic data from the GEO database and mitochondrial oxidative stress-related genes from GeneCards. Using consensus clustering, we identified 64 genes associated with mitochondrial oxidative stress in DCM and further isolated five hub genes through protein-protein interaction and machine learning techniques. These genes were analyzed for functions related to immunity, drug sensitivity, and single-cell localization. Concurrently, we collected blood samples from DCM patients to validate the hub genes' expression.
Results: The study identified five hub genes related to mitochondrial oxidative stress: VCL, ABCB1, JAK2, KDR, and NGF. Expression analysis revealed high levels of VCL, ABCB1, KDR, and NGF in the non-failing (NF) group, while JAK2 was elevated in the DCM group (p < 0.05). Diagnostic efficacy, measured by area under the curve (AUC), was significant for VCL (76.4), ABCB1 (80.1), JAK2 (68.2), KDR (78.1), and NGF (71.8). Moreover, several drugs were identified as potential regulators of these hub genes, including Topotecan, CDK9_5576, Acetalax, Afatinib, and GSK591. Notably, VCL showed increased expression in DCM patient blood samples, consistent with transcriptomic and single-cell findings.
Conclusion: This research highlights key genes associated with mitochondrial oxidative stress-VCL, ABCB1, JAK2, KDR, NGF-that show differential expression in DCM and myocardial infarction. These findings underscore their diagnostic potential and pave the way for new therapeutic strategies.
Keywords: Bioinformatics; Diagnosis; Dilated cardiomyopathy; Immune infiltration; Mitochondria; Oxidative stress.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Ethics Committee of the Suzhou Kowloon Hospital, Shanghai Jiao Tong University School of Medicine and conducted in accordance with the Declaration of Helsinki. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Exploration of dilated cardiomyopathy for biomarkers and immune microenvironment: evidence from RNA-seq.BMC Cardiovasc Disord. 2022 Jul 18;22(1):320. doi: 10.1186/s12872-022-02759-7. BMC Cardiovasc Disord. 2022. PMID: 35850644 Free PMC article.
-
Integrated microarray analysis to identify potential biomarkers and therapeutic targets in dilated cardiomyopathy.Mol Med Rep. 2020 Aug;22(2):915-925. doi: 10.3892/mmr.2020.11145. Epub 2020 May 14. Mol Med Rep. 2020. PMID: 32626989 Free PMC article.
-
Exploring the pathogenesis and immune infiltration in dilated cardiomyopathy complicated with atrial fibrillation by bioinformatics analysis.Front Immunol. 2023 Jan 17;14:1049351. doi: 10.3389/fimmu.2023.1049351. eCollection 2023. Front Immunol. 2023. PMID: 36733486 Free PMC article.
-
Precision medicine applications in dilated cardiomyopathy: Advancing personalized care.Curr Probl Cardiol. 2025 Jul;50(7):103076. doi: 10.1016/j.cpcardiol.2025.103076. Epub 2025 May 15. Curr Probl Cardiol. 2025. PMID: 40381754 Review.
-
Where genome meets phenome: rationale for integrating genetic and protein biomarkers in the diagnosis and management of dilated cardiomyopathy and heart failure.J Am Coll Cardiol. 2012 Jul 24;60(4):283-9. doi: 10.1016/j.jacc.2012.05.005. J Am Coll Cardiol. 2012. PMID: 22813604 Review.
References
-
- Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, Dubourg O, Kühl U, Maisch B, McKenna WJ, Monserrat L, Pankuweit S, Rapezzi C, Seferovic P, Tavazzi L, Keren A. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J. 2008;29:270–6. - PubMed
-
- Taylor DO, Edwards LB, Boucek MM, Trulock EP, Aurora P, Christie J, Dobbels F, Rahmel AO, Keck BM, Hertz MI. Registry of the International Society for Heart and Lung Transplantation: twenty-fourth official adult heart transplant report–2007. J Heart Lung Transplant. 2007;26:769–81. - PubMed
-
- Felker GM, Thompson RE, Hare JM, Hruban RH, Clemetson DE, Howard DL, Baughman KL, Kasper EK. Underlying causes and long-term survival in patients with initially unexplained cardiomyopathy. N Engl J Med. 2000;342:1077–84. - PubMed
-
- Weintraub RG, Semsarian C, Macdonald P. Dilated cardiomyopathy. Lancet. 2017;390:400–14. - PubMed
-
- Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–47. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

