Deep sequencing reveals distinct microRNA-mRNA signatures that differentiate pancreatic neuroendocrine tumor from non-diseased pancreas tissue
- PMID: 40217502
- PMCID: PMC11987397
- DOI: 10.1186/s12885-025-14043-w
Deep sequencing reveals distinct microRNA-mRNA signatures that differentiate pancreatic neuroendocrine tumor from non-diseased pancreas tissue
Abstract
Background: Only a limited number of biomarkers guide personalized management of pancreatic neuroendocrine tumors (PanNETs). Transcriptome profiling of microRNA (miRs) and mRNA has shown value in segregating PanNETs and identifying patients more likely to respond to treatment. Because miRs are key regulators of mRNA expression, we sought to integrate expression data from both RNA species into miR-mRNA interaction networks to advance our understanding of PanNET biology.
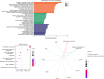
Methods: We used deep miR/mRNA sequencing on six low-grade/high-risk, well-differentiated PanNETs compared with seven non-diseased tissues to identify differentially expressed miRs/mRNAs. Then we crossed a list of differentially expressed mRNAs with a list of in silico predicted mRNA targets of the most and least abundant miRs to generate high probability miR-mRNA interaction networks.
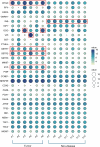
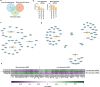
Results: Gene ontology and pathway analyses revealed several miR-mRNA pairs implicated in cellular processes and pathways suggesting perturbed neuroendocrine function (miR-7 and Reg family genes), cell adhesion (miR-216 family and NLGN1, NCAM1, and CNTN1; miR-670 and the claudins, CLDN1 and CLDN2), and metabolic processes (miR-670 and BCAT1/MPST; miR-129 and CTH).
Conclusion: These novel miR-mRNA interaction networks identified dysregulated pathways not observed when assessing mRNA alone and provide a foundation for further investigation of their utility as diagnostic and predictive biomarkers.
Keywords: Biomarkers; MicroRNA; Pancreatic neuroendocrine tumors; mRNA; miR-mRNA interaction networks.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: The study was conducted in accordance with the Declaration of Helsinki. PanNET samples were approved for research purposes through the Queen’s University Health Sciences & Affiliated Teaching Hospitals Research Ethics Board (PATH-145-14) and provided under a Material Transfer Agreement with Ontario Institute for Cancer Research. Non-diseased pancreatic tissues were obtained from the Surgical Pathology Archive in the Department of Pathology and Molecular Medicine at Queen’s University. Samples were approved for research through Research Ethics Board at Queen’s University (PATH-145-14). Written informed consent was obtained from all participants by the Ontario Institute for Cancer Research. Consent for publication: Consent for publication was obtained from all participants by the Ontario Institute for Cancer Research. Competing interests: N.M., N.V., M.G., S.B., P.C., and E.D.P. are employees of MultiplexDX. All other authors declare no competing interests.
Figures


Similar articles
-
miRNA profile in pancreatic neuroendocrine tumors: Preliminary results.Sci Prog. 2025 Jan-Mar;108(1):368504251326864. doi: 10.1177/00368504251326864. Epub 2025 Mar 28. Sci Prog. 2025. PMID: 40152231 Free PMC article.
-
A Cross-Species Analysis in Pancreatic Neuroendocrine Tumors Reveals Molecular Subtypes with Distinctive Clinical, Metastatic, Developmental, and Metabolic Characteristics.Cancer Discov. 2015 Dec;5(12):1296-313. doi: 10.1158/2159-8290.CD-15-0068. Epub 2015 Oct 7. Cancer Discov. 2015. PMID: 26446169 Free PMC article.
-
Differences between Well-Differentiated Neuroendocrine Tumors and Ductal Adenocarcinomas of the Pancreas Assessed by Multi-Omics Profiling.Int J Mol Sci. 2020 Jun 23;21(12):4470. doi: 10.3390/ijms21124470. Int J Mol Sci. 2020. PMID: 32586046 Free PMC article.
-
Diagnostic and prognostic biomarkers for pancreatic neuroendocrine neoplasms.Pathologie (Heidelb). 2024 Nov;45(Suppl 1):74-82. doi: 10.1007/s00292-024-01393-8. Epub 2024 Nov 18. Pathologie (Heidelb). 2024. PMID: 39556246 Review. English.
-
Genetics of pancreatic neuroendocrine tumors: implications for the clinic.Expert Rev Gastroenterol Hepatol. 2015;9(11):1407-19. doi: 10.1586/17474124.2015.1092383. Epub 2015 Sep 28. Expert Rev Gastroenterol Hepatol. 2015. PMID: 26413978 Free PMC article. Review.
References
-
- Parbhu SK, Adler DG. Pancreatic neuroendocrine tumors: contemporary diagnosis and management. Hosp Pract. 2016;44(3):109–19. - PubMed
-
- Li D, Rock A, Kessler J, Ballena R, Hyder S, Mo C, et al. Understanding the management and treatment of Well-Differentiated pancreatic neuroendocrine tumors: A clinician’s guide to a complex illness. JCO Oncol Pract. 2020;16(11):720–8. - PubMed
-
- Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, et al. The eighth edition AJCC cancer staging manual: continuing to build a Bridge from a population-based to a more personalized approach to cancer staging. CA Cancer J Clin. 2017;67(2):93–9. - PubMed
-
- Kos-Kudła B, Castaño JP, Denecke T, Grande E, Kjaer A, Koumarianou A, et al. European neuroendocrine tumour society (ENETS) 2023 guidance paper for nonfunctioning pancreatic neuroendocrine tumours. J Neuroendocrinol. 2023;35(12):e13343. - PubMed
-
- Perren A, Couvelard A, Scoazec JY, Costa F, Borbath I, Delle Fave G, et al. ENETS consensus guidelines for the standards of care in neuroendocrine tumors: Pathology - Diagnosis and prognostic stratification. Neuroendocrinology. 2017;105(3):196–200. - PubMed
MeSH terms
Substances
Grants and funding
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- 2019/69 MXDX-1/Ministry of Health of the Slovak Republic
- UHHK, 00179906/Ministry of Health Czech Republic
- UHHK, 00179906/Ministry of Health Czech Republic
- SVV UK, LFHK, No. 260657/Charles University, project GA UK
- SVV UK, LFHK, No. 260657/Charles University, project GA UK
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

