The influence of SARS-CoV-2 spike protein exposure on retinal development in the human retinal organoids
- PMID: 40217547
- PMCID: PMC11987193
- DOI: 10.1186/s13578-025-01383-0
The influence of SARS-CoV-2 spike protein exposure on retinal development in the human retinal organoids
Abstract
Background: Pregnant women are considered a high-risk population for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, as the virus can infect the placenta and embryos. Recently, SARS-CoV-2 has been widely reported to cause retinal pathological changes and to infect the embryonic retina. The infection of host cells by SARS-CoV-2 is primarily mediated through spike (S) protein, which also plays a crucial role in the pathogenesis of SARS-CoV-2. However, it remains poorly understood how the S protein of SARS-CoV-2 affects retinal development, and the underlying mechanism has not yet been clarified.
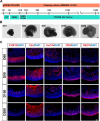
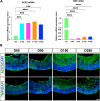
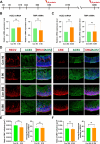
Methods: We used human embryonic stem cell-derived retinal organoids (hEROs) as a model to study the effect of S protein exposure at different stages of retinal development. hEROs were treated with 2 μg/mL of S protein on days 90 and 280. Immunofluorescence staining, RNA sequencing, and RT-PCR were performed to assess the influence of S protein exposure on retinal development at both early and late stages.
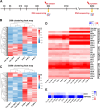
Results: The results showed that ACE2 and TMPRSS2, the receptors facilitating SARS-CoV-2 entry into host cells, were expressed in hEROs. Exposure to the S protein induced an inflammatory response in both the early and late stages of retinal development in the hEROs. Additionally, RNA sequencing indicated that early exposure of the S protein to hEROs affected nuclear components and lipid metabolism, while late-stages exposure resulted in changes to cell membrane components and the extracellular matrix.
Conclusion: This work highlights the differential effects of SARS-CoV-2 S protein exposure on retinal development at both early and late stages, providing insights into the cellular and molecular mechanisms underlying SARS-CoV-2-induced developmental impairments in the human retina.
Keywords: Retinal development; Retinal organoid; SARS-CoV-2; Spike protein.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures


Similar articles
-
Expression of Endogenous Angiotensin-Converting Enzyme 2 in Human Induced Pluripotent Stem Cell-Derived Retinal Organoids.Int J Mol Sci. 2021 Jan 28;22(3):1320. doi: 10.3390/ijms22031320. Int J Mol Sci. 2021. PMID: 33525682 Free PMC article.
-
Kidney organoids reveal redundancy in viral entry pathways during ACE2-dependent SARS-CoV-2 infection.J Virol. 2024 Mar 19;98(3):e0180223. doi: 10.1128/jvi.01802-23. Epub 2024 Feb 9. J Virol. 2024. PMID: 38334329 Free PMC article.
-
Modifications of lipid pathways restrict SARS-CoV-2 propagation in human induced pluripotent stem cell-derived 3D airway organoids.J Adv Res. 2024 Jun;60:127-140. doi: 10.1016/j.jare.2023.08.005. Epub 2023 Aug 7. J Adv Res. 2024. PMID: 37557954 Free PMC article.
-
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review.
-
Targeting the viral-entry facilitators of SARS-CoV-2 as a therapeutic strategy in COVID-19.J Med Virol. 2021 Sep;93(9):5260-5276. doi: 10.1002/jmv.27019. Epub 2021 May 3. J Med Virol. 2021. PMID: 33851732 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

