Permeable Lung Vasculature Creates Chemoresistant Endothelial Niche by Producing SERPINE1 at Breast Cancer Metastatic Sites
- PMID: 40217581
- PMCID: PMC12127090
- DOI: 10.1111/cas.70050
Permeable Lung Vasculature Creates Chemoresistant Endothelial Niche by Producing SERPINE1 at Breast Cancer Metastatic Sites
Abstract
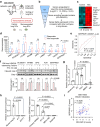
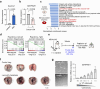
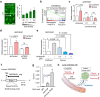
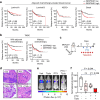
Chemotherapy resistance remains a major obstacle for eradicating metastatic cancer cells in distant organs. We identified that endothelial cells (ECs) in the lungs, where breast cancer cells often metastasize, form a chemoresistant perivascular niche for disseminated breast cancer cells. By investigating the lung EC secretome activated by metastasis, we found that serine protease inhibitor family E member 1 (SERPINE1), encoded by Serpine1, is upregulated in metastasis-associated lung ECs. This upregulation shields cancer cells from paclitaxel-induced apoptosis and promotes cancer stem cell properties. Serpine1 expression appears to be driven by YAP-TEAD activation in lung ECs that lose cell-cell contact, a phenomenon associated with increased vascular permeability in lungs affected by metastasis. Crucially, pharmacological inhibition of SERPINE1 enhances the chemotherapy sensitivity of metastatic breast cancer cells in the lung. Overall, our findings underscore the pivotal role of the vascular niche, which produces SERPINE1, in conferring chemoresistance to breast cancer cells during metastatic progression in the lungs.
Keywords: breast cancer; chemotherapy resistance; metastasis; perivascular niche; permeability.
© 2025 The Author(s). Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Prof. Noriko Gotoh is an Editorial board member of Cancer Science. Other authors do not have conflicts of interest to declare.
Figures


References
-
- Bianchini G., De Angelis C., Licata L., and Gianni L., “Treatment Landscape of Triple‐Negative Breast Cancer—Expanded Options, Evolving Needs,” Nature Reviews Clinical Oncology 19 (2022): 91–113. - PubMed
-
- de Visser K. E. and Joyce J. A., “The Evolving Tumor Microenvironment: From Cancer Initiation to Metastatic Outgrowth,” Cancer Cell 41 (2023): 374–403. - PubMed
MeSH terms
Substances
Grants and funding
- 2022A3360/Astellas Foundation for Research on Metabolic Disorders
- 22-25432/Princess Takamatsu Cancer Research Fund
- 23-255016/Princess Takamatsu Cancer Research Fund
- JP21H02761/Japan Society for the Promotion of Science
- JP23K18237/Japan Society for the Promotion of Science
- JP24K02304/Japan Society for the Promotion of Science
- JP22K20818/Japan Society for the Promotion of Science
- JP23K06630/Japan Society for the Promotion of Science
- JPMXP1323015484/Ministry of Education, Culture, Sports, Science and Technology
- JPMJSP2135/Japan Science and Technology Agency
- 202220235/The Uehara Memorial Foundation
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

