Repurposing Vancomycin as a Potential Antiviral Agent Against PEDV via nsp13 Helicase Inhibition
- PMID: 40218318
- PMCID: PMC11987754
- DOI: 10.3390/ani15070923
Repurposing Vancomycin as a Potential Antiviral Agent Against PEDV via nsp13 Helicase Inhibition
Abstract
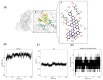
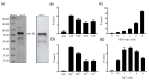
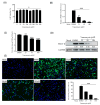
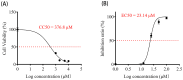
Porcine epidemic diarrhea virus (PEDV) causes a highly contagious intestinal disease with severe economic impacts on the global swine industry. The non-structural protein 13 (nsp13), a viral helicase, is essential for viral replication, making it a promising target for antiviral drug development. In this study, through virtual screening and molecular dynamics simulations, Vancomycin, a small-molecule drug also clinically used as an antibacterial agent, was identified to exhibit a stable binding affinity for PEDV nsp13. The NTPase and ATP-dependent RNA helicase activities of PEDV nsp13 were confirmed in vitro, and the optimal biochemical reaction conditions for its dsRNA unwinding activity were established. Further experiments demonstrated that Vancomycin effectively inhibited the dual enzymatic activities of PEDV nsp13 and reduced PEDV infections in vitro. This research highlights Vancomycin as a novel inhibitor of PEDV nsp13, providing valuable mechanistic insights and serving as a model for antiviral drug discovery. While this study suggests its potential for repurposing as a therapeutic agent against PEDV, further investigations are required to evaluate its feasibility in vivo, particularly in terms of safety, efficacy, and practical applicability.
Keywords: antiviral drug; helicase inhibitor; nsp13; porcine epidemic diarrhea virus; vancomycin; virtual screening.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Ferrara G., Nocera F.P., Longobardi C., Ciarcia R., Fioretti A., Damiano S., Iovane G., Pagnini U., Montagnaro S. Retrospective Serosurvey of Three Porcine Coronaviruses among the Wild Boar (Sus scrofa) Population in the Campania Region of Italy. J. Wildl. Dis. 2022;58:887–891. doi: 10.7589/JWD-D-21-00196. - DOI - PubMed
-
- Ferrara G., D’Anza E., Rossi A., Improda E., Iovane V., Pagnini U., Iovane G., Montagnaro S. A Serological Investigation of Porcine Reproductive and Respiratory Syndrome and Three Coronaviruses in the Campania Region, Southern Italy. Viruses. 2023;15:300. doi: 10.3390/v15020300. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

