Farming Practices, Biosecurity Gaps, and Genetic Insights into African Swine Fever Virus in the Iringa and Ruvuma Regions of Tanzania
- PMID: 40218400
- PMCID: PMC11987749
- DOI: 10.3390/ani15071007
Farming Practices, Biosecurity Gaps, and Genetic Insights into African Swine Fever Virus in the Iringa and Ruvuma Regions of Tanzania
Abstract
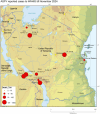
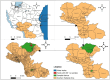
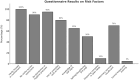
African Swine Fever Virus (ASFV) genotype II dominates outbreaks in Tanzania's Southern Highlands, continuing to persist as the dominant strain over a decade after its first incursion in 2010. A total of 205 samples from 120 holdings were collected, with 21 confirmed ASFV-positive animals from 14 holdings. Molecular analysis revealed genetic uniformity among isolates, all clustering within ASFV genotype II. Poor biosecurity measures, such as feeding of untreated swill (80% of holdings) and lack of restrictions on visitors (90% of holdings), were identified as risk factors. Additionally, co-infection with porcine circovirus-2 (PCV-2) further complicates disease management. This study underscores the urgent need for enhanced biosecurity and farmer education to mitigate ASFV outbreaks in endemic regions.
Keywords: ASFV; Tanzania; biosecurity; molecular characterization; pig farming.
Conflict of interest statement
The authors declare no conflicts of interest. The project or effort depicted was or is sponsored by the United States Department of Defense, Defense Threat Reduction Agency. The content of the information does not necessarily reflect the position or the policy of the Federal Government of the United States, and no official endorsement should be inferred. The views expressed in this publication are those of the author(s) and do not necessarily reflect the views or policies of the Food and Agriculture Organization of the United Nations.
Figures



Similar articles
-
Persistent domestic circulation of African swine fever virus in Tanzania, 2015-2017.BMC Vet Res. 2020 Oct 1;16(1):369. doi: 10.1186/s12917-020-02588-w. BMC Vet Res. 2020. PMID: 33004025 Free PMC article.
-
African swine fever: A review of cleaning and disinfection procedures in commercial pig holdings.Res Vet Sci. 2020 Oct;132:262-267. doi: 10.1016/j.rvsc.2020.06.009. Epub 2020 Jun 10. Res Vet Sci. 2020. PMID: 32693250 Review.
-
Transboundary risk of African swine fever (ASF): Detection of ASF virus genotype II in pork products carried by international travelers to Indonesia.Vet World. 2025 Feb;18(2):280-286. doi: 10.14202/vetworld.2025.280-286. Epub 2025 Feb 13. Vet World. 2025. PMID: 40182827 Free PMC article.
-
Semiquantitative Risk Evaluation Reveals Drivers of African Swine Fever Virus Transmission in Smallholder Pig Farms and Gaps in Biosecurity, Tanzania.Vet Med Int. 2024 May 13;2024:4929141. doi: 10.1155/2024/4929141. eCollection 2024. Vet Med Int. 2024. PMID: 38770528 Free PMC article.
-
African Swine Fever in Cameroon: A Review.Pathogens. 2021 Apr 1;10(4):421. doi: 10.3390/pathogens10040421. Pathogens. 2021. PMID: 33916101 Free PMC article. Review.
References
-
- Sanchez-Vizcaino J.M., Laddomada A., Arias M.L. In: Diseases of Swine. 11th ed. Zimmerman J.J., Karriker L.A., Ramirez A., Schwartz K.J., Stevenson G.W., Zhang J., editors. Volume 25. Wiley Blackwell; Hoboken, NJ, USA: 2019. pp. 443–452.
LinkOut - more resources
Full Text Sources

