A Quadruplex RT-qPCR for the Detection of Porcine Sapelovirus, Porcine Kobuvirus, Porcine Teschovirus, and Porcine Enterovirus G
- PMID: 40218401
- PMCID: PMC11987865
- DOI: 10.3390/ani15071008
A Quadruplex RT-qPCR for the Detection of Porcine Sapelovirus, Porcine Kobuvirus, Porcine Teschovirus, and Porcine Enterovirus G
Abstract
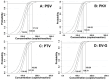
Porcine sapelovirus (PSV), porcine kobuvirus (PKV), porcine teschovirus (PTV), and porcine enterovirus G (EV-G) are all important viruses in the swine industry. These viruses play important roles in the establishment of similar clinical signs of diseases in pigs, including diarrhea, encephalitis, and reproductive and respiratory disorders. The early accurate detection of these viruses is crucial for dealing with these diseases. In order for the differential detection of these four viruses, specific primers and TaqMan probes were designed for the conserved regions in the 5' untranslated region (UTR) of these four viruses, and one-step quadruplex reverse-transcription real-time quantitative PCR (RT-qPCR) for the detection of PSV, PKV, PTV, and EV-G was developed. The results showed that this assay had the advantages of high sensitivity, strong specificity, excellent repeatability, and simple operation. Probit regression analysis showed that the assay obtained low limits of detection (LODs) for PSV, PKV, PTV, and EV-G, with 146.02, 143.83, 141.92, and 139.79 copies/reaction, respectively. The assay showed a strong specificity of detecting only PSV, PKV, PTV, and EV-G, and had no cross-reactivity with other control viruses. The assay exhibited excellent repeatability of the intra-assay coefficient of variation (CV) of 0.28-1.58% and the inter-assay CV of 0.20-1.40%. Finally, the developed quadruplex RT-qPCR was used to detect 1823 fecal samples collected in Guangxi Province, China between January 2024 and December 2024. The results indicated that the positivity rates of PSV, PKV, PTV, and EV-G were 15.25% (278/1823), 21.72% (396/1823), 18.82% (343/1823), and 27.10% (494/1823), respectively, and there existed phenomena of mixed infections. Compared with the reference RT-qPCR/RT-PCR established for these four viruses, the coincidence rates for the detection results of PSV, PKV, PTV, and EV-G reached 99.51%, 99.40%, 99.51%, and 99.01%, respectively. In conclusions, the developed quadruplex RT-qPCR could simultaneously detect PSV, PKV, PTV, and EV-G, and provided an efficient and convenient detection method to monitor the epidemic status and variation of these viruses.
Keywords: co-infection; multiplex RT-qPCR; porcine enterovirus G (EV-G); porcine kobuvirus (PKV); porcine sapelovirus (PSV); porcine teschovirus (PTV).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
First report of Porcine teschovirus (PTV), Porcine sapelovirus (PSV) and Enterovirus G (EV-G) in pig herds of Brazil.Trop Anim Health Prod. 2014 Mar;46(3):523-8. doi: 10.1007/s11250-013-0523-z. Epub 2013 Dec 22. Trop Anim Health Prod. 2014. PMID: 24362793
-
Porcine teschovirus, sapelovirus, and enterovirus in Swiss pigs: multiplex RT-PCR investigation of viral frequencies and disease association.J Vet Diagn Invest. 2021 Sep;33(5):864-874. doi: 10.1177/10406387211025827. Epub 2021 Jun 21. J Vet Diagn Invest. 2021. PMID: 34151653 Free PMC article.
-
[Clinical detection of seven porcine diarrhea-associated viruses and evolution analysis of porcine kobuvirus].Sheng Wu Gong Cheng Xue Bao. 2017 Aug 25;33(8):1292-1303. doi: 10.13345/j.cjb.170113. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 28853257 Chinese.
-
Establishment and Application of a Triplex Real-Time Reverse-Transcription Polymerase Chain Reaction Assay for Differentiation of PEDV, TGEV and PKV.Vet Sci. 2024 Sep 6;11(9):413. doi: 10.3390/vetsci11090413. Vet Sci. 2024. PMID: 39330793 Free PMC article.
-
A Systematic Review: Is Porcine Kobuvirus Causing Gastrointestinal Disease in Young Pigs?Vet Sci. 2023 Apr 11;10(4):286. doi: 10.3390/vetsci10040286. Vet Sci. 2023. PMID: 37104441 Free PMC article. Review.
Cited by
-
Colorimetric reverse transcription loop-mediated isothermal amplification assay for visual, sensitive, and specific detection of enterovirus G.BMC Vet Res. 2025 Aug 29;21(1):529. doi: 10.1186/s12917-025-04970-y. BMC Vet Res. 2025. PMID: 40883771 Free PMC article.
-
Genetic and Evolutionary Analysis of Porcine Kobuvirus in Guangxi Province, Southern China, Between 2021 and 2025.Microorganisms. 2025 Aug 17;13(8):1921. doi: 10.3390/microorganisms13081921. Microorganisms. 2025. PMID: 40871424 Free PMC article.
References
-
- Boros Á., László Z., Pankovics P., Marosi A., Albert M., Cságola A., Bíró H., Fahsbender E., Delwart E., Reuter G. High Prevalence, Genetic Diversity and a Potentially Novel Genotype of Sapelovirus A (Picornaviridae) in Enteric and Respiratory Samples in Hungarian Swine Farms. J. Gen. Virol. 2020;101:609–621. doi: 10.1099/jgv.0.001410. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

