Identification of Candidate Genes and eQTLs Related to Porcine Reproductive Function
- PMID: 40218432
- PMCID: PMC11987867
- DOI: 10.3390/ani15071038
Identification of Candidate Genes and eQTLs Related to Porcine Reproductive Function
Abstract
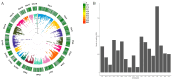
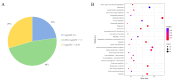
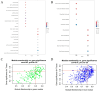
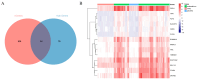
Expression quantitative trait locus (eQTL) mapping is an effective tool for identifying genetic variations that regulate gene expression. An increasing number of studies suggested that SNPs associated with complex traits in farm animals are considered as expression quantitative trait loci. Identifying eQTLs associated with gene expression levels in the endometrium helps to unravel the regulatory mechanisms of genes related to reproductive functions in this tissue and provides molecular markers for the genetic improvement of high-fertility sow breeding. In this study, 218 RNA-seq data from pig endometrial tissue were used for eQTL analysis to identify genetic variants regulating gene expression. Additionally, weighted gene co-expression network analysis (WGCNA) was performed to identify hub genes involved in reproductive functions. The eQTL analysis identified 34,876 significant cis-eQTLs regulating the expression of 5632 genes (FDR ≤ 0.05), and 90 hub genes were identified by WGCNA analysis. By integrating eQTL and WGCNA results, 14 candidate genes and 16 fine-mapped cis-eQTLs were identified, including FRK, ARMC3, SLC35F3, TMEM72, FFAR4, SOWAHA, PSPH, FMO5, HPN, FUT2, RAP1GAP, C6orf52, SEL1L3, and CLGN, which were involved in the physiological processes of reproduction in sows through hormone regulation, cell adhesion, and amino acid and lipid metabolism. These eQTLs regulate the high expression of candidate genes in the endometrium, thereby affecting reproductive-related physiological functions. These findings enhance our understanding of the genetic basis of reproductive traits and provide valuable genetic markers for marker-assisted selection (MAS), which can be applied to improve sow fecundity and optimize breeding strategies for high reproductive performance.
Keywords: WGCNA; eQTL; endometrium; pigs; reproductive function.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Tissue specific regulation of transcription in endometrium and association with disease.Hum Reprod. 2020 Feb 29;35(2):377-393. doi: 10.1093/humrep/dez279. Hum Reprod. 2020. PMID: 32103259 Free PMC article.
-
Characterization of eQTLs associated with androstenone by RNA sequencing in porcine testis.Physiol Genomics. 2019 Oct 1;51(10):488-499. doi: 10.1152/physiolgenomics.00125.2018. Epub 2019 Aug 2. Physiol Genomics. 2019. PMID: 31373884
-
The genetic regulation of transcription in human endometrial tissue.Hum Reprod. 2017 Apr 1;32(4):893-904. doi: 10.1093/humrep/dex006. Hum Reprod. 2017. PMID: 28177073
-
Genetic variations (eQTLs) in muscle transcriptome and mitochondrial genes, and trans-eQTL molecular pathways in feed efficiency from Danish breeding pigs.PLoS One. 2020 Sep 17;15(9):e0239143. doi: 10.1371/journal.pone.0239143. eCollection 2020. PLoS One. 2020. PMID: 32941478 Free PMC article.
-
Genetic approaches to the improvement of fertility traits in the pig.Vet J. 2006 Sep;172(2):234-47. doi: 10.1016/j.tvjl.2005.11.013. Epub 2006 Jan 19. Vet J. 2006. PMID: 16426876 Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

