The Riemannian Means Field Classifier for EEG-Based BCI Data
- PMID: 40218817
- PMCID: PMC11991455
- DOI: 10.3390/s25072305
The Riemannian Means Field Classifier for EEG-Based BCI Data
Abstract
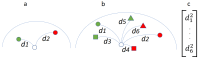
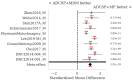
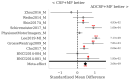
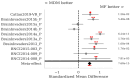
: A substantial amount of research has demonstrated the robustness and accuracy of the Riemannian minimum distance to mean (MDM) classifier for all kinds of EEG-based brain-computer interfaces (BCIs). This classifier is simple, fully deterministic, robust to noise, computationally efficient, and prone to transfer learning. Its training is very simple, requiring just the computation of a geometric mean of a symmetric positive-definite (SPD) matrix per class. We propose an improvement of the MDM involving a number of power means of SPD matrices instead of the sole geometric mean. By the analysis of 20 public databases, 10 for the motor-imagery BCI paradigm and 10 for the P300 BCI paradigm, comprising 587 individuals in total, we show that the proposed classifier clearly outperforms the MDM, approaching the state-of-the art in terms of performance while retaining the simplicity and the deterministic behavior. In order to promote reproducible research, our code will be released as open source.
Keywords: BCI; EEG; P300; Riemannian geometry; classification; motor imagery.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



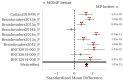


Similar articles
-
Enhancing motor imagery EEG classification with a Riemannian geometry-based spatial filtering (RSF) method.Neural Netw. 2025 Aug;188:107511. doi: 10.1016/j.neunet.2025.107511. Epub 2025 Apr 22. Neural Netw. 2025. PMID: 40294568
-
Decoding Multi-Class Motor Imagery and Motor Execution Tasks Using Riemannian Geometry Algorithms on Large EEG Datasets.Sensors (Basel). 2023 May 25;23(11):5051. doi: 10.3390/s23115051. Sensors (Basel). 2023. PMID: 37299779 Free PMC article.
-
Efficient Classification of Motor Imagery Electroencephalography Signals Using Deep Learning Methods.Sensors (Basel). 2019 Apr 11;19(7):1736. doi: 10.3390/s19071736. Sensors (Basel). 2019. PMID: 30978978 Free PMC article.
-
A review of classification algorithms for EEG-based brain-computer interfaces: a 10 year update.J Neural Eng. 2018 Jun;15(3):031005. doi: 10.1088/1741-2552/aab2f2. Epub 2018 Feb 28. J Neural Eng. 2018. PMID: 29488902 Review.
-
Riemannian Approaches in Brain-Computer Interfaces: A Review.IEEE Trans Neural Syst Rehabil Eng. 2017 Oct;25(10):1753-1762. doi: 10.1109/TNSRE.2016.2627016. Epub 2016 Nov 9. IEEE Trans Neural Syst Rehabil Eng. 2017. PMID: 27845666 Review.
References
-
- Chevallier S., Carrara I., Aristimunha B., Guetschel P., Sedlar S., Lopes B., Velut S., Khazem S., Moreau T. The largest EEG-based BCI reproducibility study for open science: The MOABB benchmark. arXiv. 2024 doi: 10.48550/arXiv.2404.15319.2404.15319 - DOI
-
- Levi-Civita T. Lezioni Di Calcolo Differenziale Assoluto. Nicola Zanichelli Editore; Bologna, Italy: 1925.
-
- Bhatia R. Positive Definite Matrices. Princeton University Press; Princeton, NJ, USA: 2015.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

