N6-Methyladenosine Modification in the Metabolic Dysfunction-Associated Steatotic Liver Disease
- PMID: 40218916
- PMCID: PMC11990428
- DOI: 10.3390/nu17071158
N6-Methyladenosine Modification in the Metabolic Dysfunction-Associated Steatotic Liver Disease
Abstract
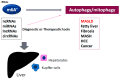
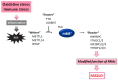
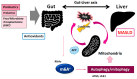
Epigenetics of N6-methyladenine (m6A) modification may play a key role during the regulation of various diseases, including metabolic dysfunction-associated steatotic liver disease (MASLD). The m6A modification has been shown to be accomplished via the exploitation of several players such as methyltransferases, demethylases, and/or methylation-binding molecules. Significantly, the m6A methylation can regulate the key eukaryotic transcriptome by affecting the subcellular localization, splicing, export, stability, translation, and decay of those RNAs. An increasing amount of data has designated that the m6A modification of RNAs can also modulate the expression of autophagy-related genes, which could also control the autophagy in hepatocytes. Oxidative stress with reactive oxygen species (ROS) can induce m6A RNA methylation, which might be associated with the regulation of mitochondrial autophagy (mitophagy) and/or the development of MASLD. Therefore, both autophagy and the m6A modification could play important roles in regulating the pathogenesis of MASLD. Comprehending the relationship between m6A and mitophagy may be helpful for the development of future therapeutic strategies against MASLD. This review would advance the understanding of the regulatory mechanisms of m6A RNA modification, focusing on the impact of mitochondrial dysregulation and mitophagy in the liver with MASLD.
Keywords: MASLD; N6-methyladenine; RNA binding protein; autophagy; liver dysfunction; mitophagy; non-coding RNA; reactive oxygen species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Hardwick J.P., Song B.J., Rote P., Leahy C., Lee Y.K., Wolf A.R., Diegisser D., Garcia V. The CYP4/20-HETE/GPR75 axis in the progression metabolic dysfunction-associated steatosis liver disease (MASLD) to chronic liver disease. Front. Physiol. 2025;15:1497297. doi: 10.3389/fphys.2024.1497297. - DOI - PMC - PubMed
-
- Alarcón-Vila C., Insausti-Urkia N., Torres S., Segalés-Rovira P., Conde de la Rosa L., Nuñez S., Fucho R., Fernández-Checa J.C., García-Ruiz C. Dietary and genetic disruption of hepatic methionine metabolism induce acid sphingomyelinase to promote steatohepatitis. Redox Biol. 2023;59:102596. doi: 10.1016/j.redox.2022.102596. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

