The Influence of an AI-Driven Personalized Nutrition Program on the Human Gut Microbiome and Its Health Implications
- PMID: 40219016
- PMCID: PMC11990151
- DOI: 10.3390/nu17071260
The Influence of an AI-Driven Personalized Nutrition Program on the Human Gut Microbiome and Its Health Implications
Abstract
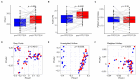
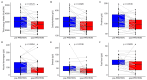
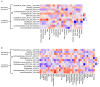
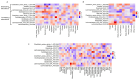
Background/Objectives: Personalized nutrition programs enhanced with artificial intelligence (AI)-based tools hold promising potential for the development of healthy and sustainable diets and for disease prevention. This study aimed to explore the impact of an AI-based personalized nutrition program on the gut microbiome of healthy individuals. Methods: An intervention using an AI-based mobile application for personalized nutrition was applied for six weeks. Fecal and blood samples from 29 healthy participants (females 52%, mean age 35 years) were collected at baseline and at six weeks. Gut microbiome through 16s ribosomal RNA (rRNA) amplicon sequencing, anthropometric and biochemical data were analyzed at both timepoints. Dietary assessment was performed using food frequency questionnaires. Results: A significant increase in richness (Chao1, 220.4 ± 58.5 vs. 241.5 ± 60.2, p = 0.024) and diversity (Faith's phylogenetic diversity, 15.5 ± 3.3 vs. 17.3 ± 2.8, p = 0.0001) was found from pre- to post-intervention. Following the intervention, the relative abundance of genera associated with the reduction in cholesterol and heart disease risk (e.g., Eubacterium coprostanoligenes group and Oscillobacter) was significantly increased, while the abundance of inflammation-associated genera (e.g., Eubacterium ruminantium group and Gastranaerophilales) was decreased. Alterations in the abundance of several butyrate-producing genera were also found (e.g., increase in Faecalibacterium, decrease in Bifidobacterium). Further, a decrease in carbohydrate (272.2 ± 97.7 vs. 222.9 ± 80.5, p = 0.003) and protein (113.6 ± 38.8 vs. 98.6 ± 32.4, p = 0.011) intake, as well as a reduction in waist circumference (78.4 ± 12.1 vs. 77.2 ± 11.2, p = 0.023), was also seen. Changes in the abundance of Oscillospiraceae_UCG_002 and Lachnospiraceae_UCG_004 were positively associated with changes in olive oil intake (Rho = 0.57, p = 0.001) and levels of triglycerides (Rho = 0.56, p = 0.001). Conclusions: This study highlights the potential for an AI-based personalized nutrition program to influence the gut microbiome. More research is now needed to establish the use of gut microbiome-informed strategies for personalized nutrition.
Keywords: artificial intelligence; gut microbiome; human health; personalized nutrition.
Conflict of interest statement
The authors declare no conflicts of interest. The research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures


References
-
- Gardner C.D., Trepanowski J.F., Del Gobbo L.C., Hauser M.E., Rigdon J., Ioannidis J.P.A., Desai M., King A.C. Effect of Low-Fat vs Low-Carbohydrate Diet on 12-Month Weight Loss in Overweight Adults and the Association with Genotype Pattern or Insulin Secretion: The DIETFITS Randomized Clinical Trial. JAMA. 2018;319:667–679. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

