Physiology and Robustness of Yeasts Exposed to Dynamic pH and Glucose Environments
- PMID: 40219637
- PMCID: PMC12152528
- DOI: 10.1002/bit.28984
Physiology and Robustness of Yeasts Exposed to Dynamic pH and Glucose Environments
Abstract
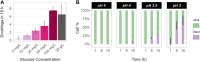
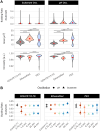
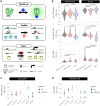
Gradients negatively affect performance in large-scale bioreactors; however, they are difficult to predict at laboratory scale. Dynamic microfluidics single-cell cultivation (dMSCC) has emerged as an important tool for investigating cell behavior in rapidly changing environments. In the present study, dMSCC, biosensors of intracellular parameters, and robustness quantification were employed to investigate the physiological response of three Saccharomyces cerevisiae strains to substrate and pH changes every 0.75-48 min. All strains showed higher sensitivity to substrate than pH oscillations. Strain-specific intracellular responses included higher relative glycolytic flux and oxidative stress response for strains PE2 and CEN.PK113-7D, respectively. Instead, the Ethanol Red strain displayed the least heterogeneous populations and the highest robustness for multiple functions when exposed to substrate oscillations. This result could arise from a positive trade-off between ATP levels and ATP stability over time. The present study demonstrates the importance of coupling physiological responses to dynamic environments with simultaneous characterization of strains, conditions, individual regimes, and robustness analysis. All these tools are a suitable add-on to traditional evaluation and screening workflows at both laboratory and industrial scale, and can help bridge the gap between these two.
Keywords: ATP; Saccharomyces cerevisiae; bioprocess; biosensors; dynamic environment; glycolytic flux; microfluidics; oxidative stress; robustness.
© 2025 The Author(s). Biotechnology and Bioengineering published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Quantifying microbial robustness in dynamic environments using microfluidic single-cell cultivation.Microb Cell Fact. 2024 Feb 9;23(1):44. doi: 10.1186/s12934-024-02318-z. Microb Cell Fact. 2024. PMID: 38336674 Free PMC article.
-
Physiology of the fuel ethanol strain Saccharomyces cerevisiae PE-2 at low pH indicates a context-dependent performance relevant for industrial applications.FEMS Yeast Res. 2014 Dec;14(8):1196-205. doi: 10.1111/1567-1364.12217. Epub 2014 Oct 31. FEMS Yeast Res. 2014. PMID: 25263709
-
Microfluidic single-cell scale-down bioreactors: A proof-of-concept for the growth of Corynebacterium glutamicum at oscillating pH values.Biotechnol Bioeng. 2022 Nov;119(11):3194-3209. doi: 10.1002/bit.28208. Epub 2022 Sep 2. Biotechnol Bioeng. 2022. PMID: 35950295
-
Four ways of implementing robustness quantification in strain characterisation.Biotechnol Biofuels Bioprod. 2023 Dec 19;16(1):195. doi: 10.1186/s13068-023-02445-6. Biotechnol Biofuels Bioprod. 2023. PMID: 38115067 Free PMC article.
-
Real-Time Monitoring of the Yeast Intracellular State During Bioprocesses With a Toolbox of Biosensors.Front Microbiol. 2022 Jan 7;12:802169. doi: 10.3389/fmicb.2021.802169. eCollection 2021. Front Microbiol. 2022. PMID: 35069506 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

