A specific pluripotency-associated eRNA controls Nanog locus by shaping the epigenetic landscape and stabilizing enhancer-promoter interaction
- PMID: 40219964
- PMCID: PMC11992674
- DOI: 10.1093/nar/gkaf274
A specific pluripotency-associated eRNA controls Nanog locus by shaping the epigenetic landscape and stabilizing enhancer-promoter interaction
Abstract
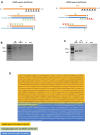
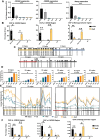
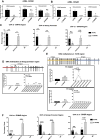
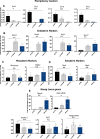
Despite a plethora of studies exploring the transcriptional regulation of the Nanog gene, the role of the enhancer RNAs (eRNAs) derived from Nanog-interacting super-enhancers (SEs) remains under-investigated. In the present study, we examined the functional role of the eRNAs transcribed from the -5 kb Nanog SE in mouse embryonic stem cells (mESCs) and found that an eRNA, here defined as -5KNAR, was essential to maintain the Nanog locus in an epigenetically active configuration, thereby ensuring pluripotency. We found that the here identified -5KNAR functionally interacts with the RAD21 protein, suggesting a role in stabilizing a cohesin complex at the Nanog locus, ensuring the generation and maintenance of an enhancer-promoter loop. Silencing of -5KNAR caused a cascade of events, including the generation of a DNA methylation wave (likely spreading from a single methylated CpG site), substantial chromatin remodeling, and loss of the enhancer-promoter loop, inducing Nanog silencing and mESC differentiation. Under these conditions, exogenous re-expression of Nanog was unable to restore either the endogenous Nanog expression or the enhancer-promoter interaction, suggesting that, at hierarchical level, the expression of the -5KNAR plays a prominent role in maintaining the pluripotency in mESCs.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures


Similar articles
-
Super-Enhancers at the Nanog Locus Differentially Regulate Neighboring Pluripotency-Associated Genes.Cell Rep. 2016 Sep 27;17(1):19-28. doi: 10.1016/j.celrep.2016.09.002. Cell Rep. 2016. PMID: 27681417 Free PMC article.
-
Nanog Expression in Embryonic Stem Cells - An Ideal Model System to Dissect Enhancer Function.Bioessays. 2017 Dec;39(12):10.1002/bies.201700086. doi: 10.1002/bies.201700086. Epub 2017 Oct 4. Bioessays. 2017. PMID: 28977693 Free PMC article. Review.
-
Adequate concentration of B cell leukemia/lymphoma 3 (Bcl3) is required for pluripotency and self-renewal of mouse embryonic stem cells via downregulation of Nanog transcription.BMB Rep. 2018 Feb;51(2):92-97. doi: 10.5483/bmbrep.2018.51.2.219. BMB Rep. 2018. PMID: 29335071 Free PMC article.
-
Long-Range Enhancer Interactions Are Prevalent in Mouse Embryonic Stem Cells and Are Reorganized upon Pluripotent State Transition.Cell Rep. 2018 Mar 6;22(10):2615-2627. doi: 10.1016/j.celrep.2018.02.040. Cell Rep. 2018. PMID: 29514091 Free PMC article.
-
Mechanisms of pluripotency maintenance in mouse embryonic stem cells.Cell Mol Life Sci. 2017 May;74(10):1805-1817. doi: 10.1007/s00018-016-2438-0. Epub 2016 Dec 20. Cell Mol Life Sci. 2017. PMID: 27999898 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

