Nuclear AGO2 supports influenza A virus replication through type-I interferon regulation
- PMID: 40219968
- PMCID: PMC11992678
- DOI: 10.1093/nar/gkaf268
Nuclear AGO2 supports influenza A virus replication through type-I interferon regulation
Abstract
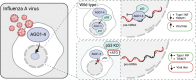
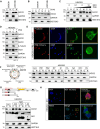
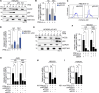
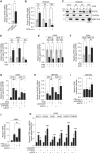
The role of Argonaute (AGO) proteins and the RNA interference (RNAi) machinery in mammalian antiviral response has been debated. Therefore, we set out to investigate how mammalian RNAi impacts influenza A virus (IAV) infection. We reveal that IAV infection triggers nuclear accumulation of AGO2, which is directly facilitated by p53 activation. Mechanistically, we show that IAV induces nuclear AGO2 targeting of TRIM71and type-I interferon-pathway genes for silencing. Accordingly, Tp53-/- mice do not accumulate nuclear AGO2 and demonstrate decreased susceptibility to IAV infection. Hence, the RNAi machinery is highjacked by the virus to evade the immune system and support viral replication. Furthermore, the FDA-approved drug, arsenic trioxide, prevents p53 nuclear translocation, increases interferon response and decreases viral replication in vitro and in a mouse model in vivo. Our data indicate that targeting the AGO2:p53-mediated silencing of innate immunity may offer a promising strategy to mitigate viral infections.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

