Structural and biochemical characterization of the 3'-5' tRNA splicing ligases
- PMID: 40220997
- PMCID: PMC12135372
- DOI: 10.1016/j.jbc.2025.108506
Structural and biochemical characterization of the 3'-5' tRNA splicing ligases
Abstract
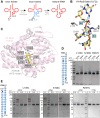
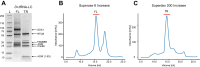
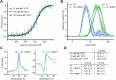
In archaea and metazoa, tRNA exons are ligated by the RNA ligases RtcB and RTCB, respectively. The metazoan RTCB forms a stable complex with four additional subunits, DDX1, FAM98B, CGI99, and ASHWIN. The role and assembly of these four components remain elusive. Furthermore, we lack structural information of how RNA substrates are recognized by 3'-5' tRNA ligases. Here, we use thiol-based chemical crosslinking to confirm the involvement of specific residues of RtcB in RNA binding, and we present a cryo-EM structure of the purified five-subunit Danio rerio tRNA ligase complex. The structure implies that the DDX1 helicase module is mobile and can modulate the activity of RTCB. Taken together, the presented results enhance our mechanistic understanding of RNA binding by 3'-5' tRNA splicing ligases and architecture of the metazoan tRNA ligase complex.
Keywords: RNA splicing; RNA-binding protein; RTCB; pre-tRNA splicing; protein complex; protein structure; protein–nucleic acid interaction; structural biology; tRNA; tRNA ligase complex.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

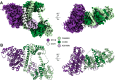

References
-
- Peebles C.L., Ogden R.C., Knapp G. Splicing of yeast tRNA precursors: a two-stage reaction. Cell. 1979;18:27–35. - PubMed
-
- Hopper A.K., Banks F., Evangelidis V. A yeast mutant which accumulates precursor tRNAs. Cell. 1978;14:211–219. - PubMed
-
- Schmidt C.A., Matera A.G. tRNA introns: presence, processing, and purpose. WIREs RNA. 2020;11 - PubMed
-
- Peebles C.L., Gegenheimer P., Abelson J. Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease. Cell. 1983;32:525–536. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

