Derivation of human trophoblast stem cells from placentas at birth
- PMID: 40221000
- PMCID: PMC12212129
- DOI: 10.1016/j.jbc.2025.108505
Derivation of human trophoblast stem cells from placentas at birth
Abstract
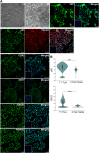
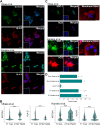
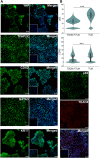
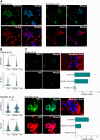
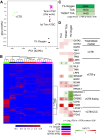
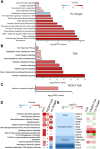
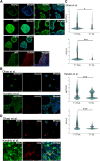
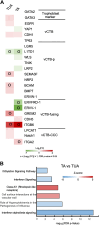
Human trophoblast stem cells (hTSCs) have emerged as a powerful tool for modeling the placental cytotrophoblast (CTB) in vitro. hTSCs were originally derived from CTBs of the first-trimester placenta or blastocyst-stage embryos in trophoblast stem cell medium (TSCM) that contains epidermal growth factor, the glycogen synthase kinase-beta inhibitor CHIR99021, the transforming growth factor-beta inhibitors A83-01 and SB431542, valproic acid, and the Rho-associated protein kinase inhibitor Y-27632. Here, we show that hTSCs can be derived from CTBs isolated from the term placenta, using TSCM supplemented with a low concentration of mitochondrial pyruvate uptake inhibitor UK5099 and lipid-rich albumin (TUA medium). Notably, hTSCs could not be derived from term CTBs using TSCM alone, or in the absence of either UK5099 or lipid-rich albumin. Strikingly, hTSCs cultured in TUA medium for a few passages could be transitioned into TSCM and cultured thereafter in TSCM. hTSCs from term CTBs cultured in TUA medium as well as those transitioned into and cultured in TSCM thereafter could be differentiated to the extravillous trophoblast and syncytiotrophoblast lineages and exhibited high transcriptome similarity with hTSCs derived from first-trimester CTBs. We anticipate that these results will enable facile derivation of hTSCs from normal and pathological placentas at birth with diverse genetic backgrounds and facilitate in vitro mechanistic studies in trophoblast biology.
Keywords: cytotrophoblast; extravillous trophoblast; human trophoblast stem cells; placenta; syncytiotrophoblast.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures

Update of
-
Derivation of human trophoblast stem cells from placentas at birth.bioRxiv [Preprint]. 2025 Feb 28:2024.05.01.592064. doi: 10.1101/2024.05.01.592064. bioRxiv. 2025. Update in: J Biol Chem. 2025 Jun;301(6):108505. doi: 10.1016/j.jbc.2025.108505. PMID: 38746283 Free PMC article. Updated. Preprint.
Similar articles
-
Derivation of human trophoblast stem cells from placentas at birth.bioRxiv [Preprint]. 2025 Feb 28:2024.05.01.592064. doi: 10.1101/2024.05.01.592064. bioRxiv. 2025. Update in: J Biol Chem. 2025 Jun;301(6):108505. doi: 10.1016/j.jbc.2025.108505. PMID: 38746283 Free PMC article. Updated. Preprint.
-
Derivation of trophoblast stem cells from naïve human pluripotent stem cells.Elife. 2020 Feb 12;9:e52504. doi: 10.7554/eLife.52504. Elife. 2020. PMID: 32048992 Free PMC article.
-
A roadmap towards targeted differentiation of mouse trophoblast stem cells into cell types of the feto-maternal exchange surface.Stem Cell Res Ther. 2025 Jul 15;16(1):371. doi: 10.1186/s13287-025-04487-z. Stem Cell Res Ther. 2025. PMID: 40660370 Free PMC article.
-
Stem cell insights into human trophoblast lineage differentiation.Hum Reprod Update. 2016 Dec;23(1):77-103. doi: 10.1093/humupd/dmw026. Epub 2016 Sep 2. Hum Reprod Update. 2016. PMID: 27591247
-
Ethical considerations for advancing research using organoid models derived from the placenta.Hum Reprod Update. 2025 Jul 1;31(4):392-401. doi: 10.1093/humupd/dmaf007. Hum Reprod Update. 2025. PMID: 40096642 Free PMC article. Review.
Cited by
-
The human placenta and its role in reproductive outcomes revisited.Physiol Rev. 2025 Oct 1;105(4):2305-2376. doi: 10.1152/physrev.00039.2024. Epub 2025 Jun 11. Physiol Rev. 2025. PMID: 40497429 Review.
References
-
- James J.L. Stem cells and pregnancy disorders: from pathological mechanisms to therapeutic horizons. Semin. Reprod. Med. 2016;34:17–26. - PubMed
-
- Goldman-Wohl D., Yagel S. Regulation of trophoblast invasion: from normal implantation to pre-eclampsia. Mol. Cell Endocrinol. 2002;187:233–238. - PubMed
-
- Chavatte-Palmer P., Tarrade A. Placentation in different mammalian species. Ann. Endocrinol (Paris). 2016;77:67–74. - PubMed
-
- Okae H., Toh H., Sato T., Hiura H., Takahashi S., Shirane K., et al. Derivation of human trophoblast stem cells. Cell Stem Cell. 2018;22:50–63.e6. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

