Predicting interval from diagnosis to delivery in preeclampsia using electronic health records
- PMID: 40221413
- PMCID: PMC11993686
- DOI: 10.1038/s41467-025-58437-7
Predicting interval from diagnosis to delivery in preeclampsia using electronic health records
Abstract
Preeclampsia is a major cause of maternal and perinatal mortality with no known cure. Delivery timing is critical to balancing maternal and fetal risks. We develop and externally validate PEDeliveryTime, a class of clinically informative models which resulted from deep-learning models, to predict the time from PE diagnosis to delivery using electronic health records. We build the models on 1533 PE cases from the University of Michigan and validate it on 2172 preeclampsia cases from the University of Florida. PEDeliveryTime full model contains only 12 features yet achieves high c-index of 0.79 and 0.74 on the Michigan and Florida data set respectively. For the early-onset preeclampsia subset, the full model reaches 0.76 and 0.67 on the Michigan and Florida test sets. Collectively, these models perform an early assessment of delivery urgency and might help to better prioritize medical resources.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


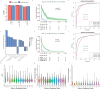


References
-
- Ives, C. W., Sinkey, R., Rajapreyar, I., Tita, A. T. N. & Oparil, S. Preeclampsia—pathophysiology and clinical presentations. J. Am. Coll. Cardiol.76, 1690–1702 (2020). - PubMed
-
- US Preventive Services Task Force, et al. Screening for preeclampsia: US preventive services task force recommendation statement. JAMA317, 1661 (2017). - PubMed
-
- Sibai, B. M. Evaluation and management of severe preeclampsia before 34 weeks’ gestation. Am. J. Obstet. Gynecol.205, 191–198 (2011). - PubMed
-
- von Dadelszen, P., Magee, L. A. & Roberts, J. M. Subclassification of Preeclampsia. Hypertension Pregnancy22, 143–148 (2003). - PubMed
MeSH terms
Grants and funding
- R01LM012373/U.S. Department of Health & Human Services | NIH | U.S. National Library of Medicine (NLM)
- R01HD084633/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R01 LM012907/LM/NLM NIH HHS/United States
- R01 LM012373/LM/NLM NIH HHS/United States
- T32GM141746/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- UL1 TR001427/TR/NCATS NIH HHS/United States
- F30 HL172988/HL/NHLBI NIH HHS/United States
- R01 HD084633/HD/NICHD NIH HHS/United States
- T32 GM141746/GM/NIGMS NIH HHS/United States
- K01 DK115632/DK/NIDDK NIH HHS/United States
- R01LM012907/U.S. Department of Health & Human Services | NIH | U.S. National Library of Medicine (NLM)
- TL1 TR001428/TR/NCATS NIH HHS/United States
- K01DK115632/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
LinkOut - more resources
Full Text Sources
Medical

