TRUSTED: The Paired 3D Transabdominal Ultrasound and CT Human Data for Kidney Segmentation and Registration Research
- PMID: 40221416
- PMCID: PMC11993632
- DOI: 10.1038/s41597-025-04467-1
TRUSTED: The Paired 3D Transabdominal Ultrasound and CT Human Data for Kidney Segmentation and Registration Research
Abstract
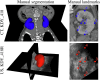
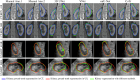
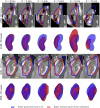
Inter-modal image registration (IMIR) and image segmentation with abdominal Ultrasound (US) data have many important clinical applications, including image-guided surgery, automatic organ measurement, and robotic navigation. However, research is severely limited by the lack of public datasets. We propose TRUSTED (the Tridimensional Renal Ultra Sound TomodEnsitometrie Dataset), comprising paired transabdominal 3DUS and CT kidney images from 48 human patients (96 kidneys), including segmentation, and anatomical landmark annotations by two experienced radiographers. Inter-rater segmentation agreement was over 93% (Dice score), and gold-standard segmentations were generated using the STAPLE algorithm. Seven anatomical landmarks were annotated, for IMIR systems development and evaluation. To validate the dataset's utility, 4 competitive Deep-Learning models for kidney segmentation were benchmarked, yielding average DICE scores from 79.63% to 90.09% for CT, and 70.51% to 80.70% for US images. Four IMIR methods were benchmarked, and Coherent Point Drift performed best with an average Target Registration Error of 4.47 mm and Dice score of 84.10%. The TRUSTED dataset may be used freely to develop and validate segmentation and IMIR methods.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Leroy, A. et al. Percutaneous renal puncture, requirements and preliminary results. arXiv preprint physics/0610209 (2006).
-
- Arun, K. S., Huang, T. S. & Blostein, S. D. Least-squares fitting of two 3-d point sets. IEEE Transactions on Pattern Analysis and Machine IntelligencePAMI-9, 698–700, 10.1109/TPAMI.1987.4767965 (1987). - PubMed
-
- Agamennoni, G., Fontana, S., Siegwart, R. Y. & Sorrenti, D. G. Point clouds registration with probabilistic data association. In 2016 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), 4092–4098, 10.1109/IROS.2016.7759602 (2016).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

