Development of a reliable risk prognostic model for lung adenocarcinoma based on the genes related to endotheliocyte senescence
- PMID: 40221448
- PMCID: PMC11993614
- DOI: 10.1038/s41598-025-95551-4
Development of a reliable risk prognostic model for lung adenocarcinoma based on the genes related to endotheliocyte senescence
Abstract
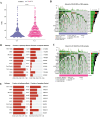
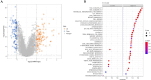
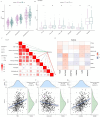
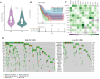
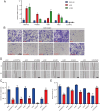
Cellular senescence is a hallmark for cancers, particularly in lung adenocarcinoma (LUAD). This study developed a risk model using senescence signature genes for LUAD patients. Based on the RNA-seq, clinical information and mutation data of LUAD patients collected from the TCGA and GEO database, we obtained 102 endotheliocyte senescence-related genes. The "ConsensusClusterPlus" R package was employed for unsupervised cluster analysis, and the "limma" was used for the differentially expressed gene (DEG) analysis. A prognosis model was created by univariate and multivariate Cox regression analysis combined with Lasso regression utilizing the "survival" and "glmnet" packages. KM survival and receiver operator characteristic curve analyses were conducted applying the "survival" and "timeROC" packages. "MCPcounter" package was used for immune infiltration analysis. Immunotherapy response analysis was performed based on the IMvigor210 and GSE78220 cohort, and drug sensitivity was predicted by the "pRRophetic" package. Cell invasion and migration were tested by carrying out Transwell and wound healing assays. According to the results, a total of 32 genes related to endotheliocyte senescence were screened to assign patients into C1 and C2 subtypes. The C2 subtype showed a significantly worse prognosis and an overall higher somatic mutation frequency, which was associated with increased activation of cancer pathways, including Myc_targets2 and angiogenesis. Then, based on the DEGs between the two subtypes, we constructed a five-gene RiskScore model with a strong classification effectiveness for short- and long-term OS prediction. High- and low-risk groups of LUAD patients were classified by the RiskScore. High-risk patients, characterized by lower immune infiltration, had poorer outcomes in both training and validation datasets. The RiskScore was associated with the immunotherapy response in LUAD. Finally, we found that potential drugs such as Cisplatin can benefit high-risk LUAD patients. In-vitro experiments demonstrated that silencing of Angiopoietin-like 4 (ANGPTL4), Gap Junction Protein Beta 3 (GJB3), Family with sequence similarity 83-member A (FAM83A), and Anillin (ANLN) reduced the number of invasive cells and the wound healing rate, while silencing of solute carrier family 34 member 2 (SLC34A2) had the opposite effect. This study, collectively speaking, developed a prognosis model with senescence signature genes to facilitate the diagnosis and treatment of LUAD.
Keywords: ConsensusClusterPlus; Immune-infiltration; Lung adenocarcinoma (LUAD); RiskScore; Senescence signature.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Tang, Z., Wang, L., Wu, G., Qin, L.& Tan, Y. FGD5 as a novel prognostic biomarker and its association with immune infiltrates in lung adenocarcinoma. Biocell47 (11), 2503–2516 (2023). - DOI
-
- Cao, H. et al. Identification of prognostic molecular subtypes and model based on CD8 + T cells for lung adenocarcinoma. Biocell48 (3), 473–490 (2024). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

