Study of coronavirus diversity in wildlife in Northern Cambodia suggests continuous circulation of SARS-CoV-2-related viruses in bats
- PMID: 40221475
- PMCID: PMC11993651
- DOI: 10.1038/s41598-025-92475-x
Study of coronavirus diversity in wildlife in Northern Cambodia suggests continuous circulation of SARS-CoV-2-related viruses in bats
Abstract
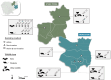
Since SARS-CoV-2's emergence, studies in Southeast Asia, including Cambodia, have identified related coronaviruses (CoVs) in rhinolophid bats. This pilot study investigates the prevalence and diversity of CoVs in wildlife from two Cambodian provinces known for wildlife trade and environmental changes, factors favoring zoonotic spillover risk. Samples were collected from 2020 to 2022 using active (capture and swabbing of bats and rodents) and non-invasive (collection of feces from bat caves and wildlife habitats) methods. RNA was screened for CoVs using conventional pan-CoVs and real-time Sarbecovirus-specific PCR systems. Positive samples were sequenced and phylogenetic analysis was performed on the partial RdRp gene. A total of 2608 samples were collected: 867 rectal swabs from bats, 159 from rodents, 41 from other wild animals, and 1541 fecal samples. The overall prevalence of CoVs was 2.0%, with a 3.3% positive rate in bats, 2.5% in rodents, and no CoVs detected in other wildlife species. Alpha-CoVs were exclusive to bats, while Beta-CoVs were found in both bats and rodents. Seven SARS-CoV-2-related viruses were identified in Rhinolophus shameli bats sampled in August 2020, March 2021, and December 2021. Our results highlight diverse CoVs in Cambodian bats and rodents and emphasize bats as significant reservoirs. They also suggest continuous circulation of bat SARS-CoV-2-related viruses may occur in a region where ecological and human factors could favor virus emergence. Continuous surveillance and integrated approaches are crucial to managing and mitigating emerging zoonotic diseases.
Keywords: Bat; Cambodia; Coronavirus; Sarbecovirus; Wildlife.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- Lau, S. K. P. et al. Genetic characterization of betacoronavirus lineage C viruses in Bats reveals marked sequence divergence in the Spike protein of pipistrellus Bat coronavirus HKU5 in Japanese pipistrelle: implications for the origin of the novel middle East respiratory syndrome coronavirus. J. Virol.87 (15), 8638–8650 (2013). - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

