Population genetic structure and demographic history of Dermacentor marginatus Sulzer, 1776 in Anatolia
- PMID: 40221486
- PMCID: PMC11993584
- DOI: 10.1038/s41598-025-97658-0
Population genetic structure and demographic history of Dermacentor marginatus Sulzer, 1776 in Anatolia
Abstract
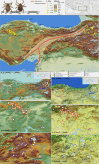
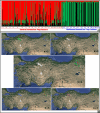
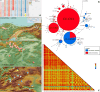
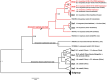
Dermacentor marginatus is a medically important tick species due to its preference humans and domestic animals as hosts and its vectorial competence, yet it remains understudied in many regions. This study aimed to examine the population structure and demographic history of D. marginatus using the cox1 and ITS2 genes, focusing on populations from Central and Northeast Anatolia-two regions on either side of the Anatolian Diagonal, a natural biogeographical barrier. A total of 361 host-seeking adult D. marginatus ticks from 31 sampling sites were analyzed, revealing 131 haplotypes for cox1 and 104 genotypes for ITS2. Neutrality tests and mismatch distribution patterns rejected the null hypothesis of the neutral theory, indicating that the population of D. marginatus in Anatolia has undergone a recent demographic expansion. Significant genetic differentiation and population structuring were observed between the Central and Northeastern Anatolian populations of D. marginatus, correlating with geographic distance and suggesting that the Anatolian Diagonal acts as a potential barrier to gene flow. Intrapopulation gene flow was higher in Central Anatolian populations compared to Northeastern Anatolian populations. Bayesian phylogeny revealed a highly divergent D. marginatus haplotype within the Northeastern Anatolian population, clustering into a Central Asian clade. Additionally, phylogenetic trees of the subgenus Serdjukovia revealed taxonomic ambiguities, including the absence of a distinct clade for D. niveus and potential misidentifications of D. marginatus and D. raskemensis specimens. Furthermore, the monophyletic relationship between D. marginatus and D. raskemensis supports the likelihood of sympatric speciation. These findings enhance our understanding of the genetic structure, phylogeography, and evolutionary dynamics of D. marginatus while providing a framework for future research on tick populations.
Keywords: Dermacentor marginatus complex; Serdjukovia; cox1; ITS2; Phylogeography; Tick evolution.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Genetic diversity analysis of Dermacentor nuttalli within Inner Mongolia, China.Parasit Vectors. 2021 Mar 1;14(1):131. doi: 10.1186/s13071-021-04625-5. Parasit Vectors. 2021. PMID: 33648549 Free PMC article.
-
Molecular characterization of Ribosomal DNA (ITS2) of hard ticks in Iran: understanding the conspecificity of Dermacentor marginatus and D. niveus.BMC Res Notes. 2020 Oct 9;13(1):478. doi: 10.1186/s13104-020-05326-5. BMC Res Notes. 2020. PMID: 33036651 Free PMC article.
-
Notes on Dermacentor ticks: redescription of D. marginatus with the synonymies of D. niveus and D. daghestanicus (Acari: Ixodidae).J Med Entomol. 1991 Jan;28(1):2-15. J Med Entomol. 1991. PMID: 2033615
-
Genetic evidence for conspecificity between Dermacentor marginatus and Dermacentor niveus.Parasitol Res. 2009 Oct;105(4):1125-32. doi: 10.1007/s00436-009-1532-8. Epub 2009 Jun 27. Parasitol Res. 2009. PMID: 19562378
-
Dermacentor everestianus Hirst, 1926 (Acari: Ixodidae): phylogenetic status inferred from molecular characteristics.Parasitol Res. 2014 Oct;113(10):3773-9. doi: 10.1007/s00436-014-4043-1. Epub 2014 Jul 23. Parasitol Res. 2014. PMID: 25049051
References
-
- Araya-Anchetta, A., Busch, J. D., Scoles, G. A. & Wagner, D. M. Thirty years of tick population genetics: A comprehensive review. Infect. Genet. Evol.29, 164–179 (2015). - PubMed
-
- De La Fuente, J. et al. Characterization of genetic diversity in Dermacentor andersoni (Acari: Ixodidae) with body size and weight polymorphism. Exp. Parasitol.109, 16–26 (2005). - PubMed
-
- Beati, L. & Klompen, H. Phylogeography of ticks (Acari: Ixodida). Annu. Rev. Entomol.64, 379–397 (2019). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

