Small molecules direct the generation of ameloblast-like cells from human embryonic stem cells
- PMID: 40221796
- PMCID: PMC11993985
- DOI: 10.1186/s13287-025-04294-6
Small molecules direct the generation of ameloblast-like cells from human embryonic stem cells
Abstract
Background: Ameloblasts present a promising avenue for the investigation of enamel and tooth regeneration. Previous protocols for directing the differentiation of human embryonic stem cells (hESCs) into dental epithelial (DE) cells involving the need for additional cells, conditional medium, and the use of costly cytokines. Importantly, ameloblasts have not been generated from hESCs in previous studies. Hence, we aimed to identify defined differentiation conditions that would solely utilize small molecules to achieve the production of ameloblasts.
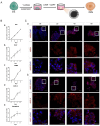
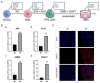
Methods: We developed a three-step strategy entailing the progression of hESCs through non-neural ectoderm (NNE) and DE to generate functional ameloblasts in vitro. Initially, the NNE fate was induced from hESCs using a 6-day differentiation protocol with 1 µmol/L Retinoic acid (RA). Subsequently, the NNE lineage was differentiated into DE by employing a combination of 1 µmol/L LDN193189 (a BMP signaling inhibitor) and 1 µmol/L XAV939 (a WNT signaling inhibitor). In the final phase, 3 µmol/L CHIR99021 (a WNT signaling activator) and 2 µmol/L DAPT (a NOTCH signaling inhibitor) were utilized to achieve the fate of ameloblasts from DE cells. Three-dimensional cultures were investigated to enhance the ameloblast differentiation ability of the induced DE cells. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) and immunofluorescence were conducted to assess the expression of lineage-specific markers. Alizarin Red S (ARS) staining was performed to evaluate the formation of mineralization nodules.
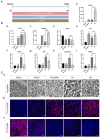
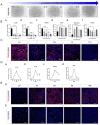
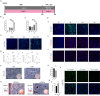
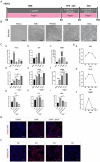
Results: The application of RA facilitated the efficient generation of NNE within a six-day period. Subsequently, upon stimulation with LDN193189 and XAV939, a notable emergence of DE cells was observed on the eighth days. By the tenth day, ameloblast-like cells derived from hESCs were generated. Upon cultivation in spheroids, these cells exhibited elevated levels of ameloblast markers AMBN and AMELX expression, suggesting that spheroid culture augments the differentiation of ameloblasts.
Conclusion: We established an efficient small molecule-based method to differentiate hESCs into ameloblast-like cells through the concerted modulation of RA, BMP, WNT, and NOTCH signaling pathways, potentially advancing research in enamel and tooth regeneration.
Keywords: Ameloblasts; Human embryonic stem cell; Small molecules; Spheroids.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: hESC cell line H9 was kindly provided by Cellapybio Biotechnology Co., LTD, they obtained the cell line from WiCell Research Institute (Madison, WI, USA) (Thomson, J. A. ``Embryonic Stem Cell Lines Derived from Human Blastocysts.`` Science 282.5391 (1998): 1145 − 147.). All healthy dental pulp tissues used for hDPSCs isolation were donated with written consent under approval by the Stomatological Hospital, College of Medicine, Xi’an Jiaotong University, China (The approved project titled “The Study of Small Molecule Compounds Inducing Differentiation of Human Pluripotent Stem Cells to Odontogenic-related Cells” was under approval by the Stomatological Hospital, College of Medicine, Xi’an Jiaotong University on Nov. 20th, 2024. The approval number is KY-GXB-20240011). Consent for publication: All authors confirm their consent for publication, if this manuscript is accepted. Conflict of interest: The authors deny any conflicts of interest with this article.
Figures
Similar articles
-
Small Molecules Promote the Rapid Generation of Dental Epithelial Cells from Human-Induced Pluripotent Stem Cells.Int J Mol Sci. 2024 Apr 9;25(8):4138. doi: 10.3390/ijms25084138. Int J Mol Sci. 2024. PMID: 38673725 Free PMC article.
-
CHIR99021 combined with retinoic acid promotes the differentiation of primordial germ cells from human embryonic stem cells.Oncotarget. 2017 Jan 31;8(5):7814-7826. doi: 10.18632/oncotarget.13958. Oncotarget. 2017. PMID: 27999196 Free PMC article.
-
Stage-Specific Role of Amelx Activation in Stepwise Ameloblast Induction from Mouse Induced Pluripotent Stem Cells.Int J Mol Sci. 2021 Jul 3;22(13):7195. doi: 10.3390/ijms22137195. Int J Mol Sci. 2021. PMID: 34281250 Free PMC article.
-
From Pluripotent Stem Cells to Organoids and Bioprinting: Recent Advances in Dental Epithelium and Ameloblast Models to Study Tooth Biology and Regeneration.Stem Cell Rev Rep. 2024 Jul;20(5):1184-1199. doi: 10.1007/s12015-024-10702-w. Epub 2024 Mar 18. Stem Cell Rev Rep. 2024. PMID: 38498295 Free PMC article. Review.
-
Essential roles of ameloblastin in maintaining ameloblast differentiation and enamel formation.Cells Tissues Organs. 2005;181(3-4):189-95. doi: 10.1159/000091380. Cells Tissues Organs. 2005. PMID: 16612084 Review.
References
-
- Mohammadi Amirabad L, Zarrintaj P, Lindemuth A, et al. Whole tooth engineering. In: Tayebi L, editor. Applications of biomedical engineering in dentistry. Cham: Springer International Publishing; 2020. pp. 443–62.
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–7. 10.1126/science.282.5391.1145. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

