Multimodal fusion of radio-pathology and proteogenomics identify integrated glioma subtypes with prognostic and therapeutic opportunities
- PMID: 40222975
- PMCID: PMC11994800
- DOI: 10.1038/s41467-025-58675-9
Multimodal fusion of radio-pathology and proteogenomics identify integrated glioma subtypes with prognostic and therapeutic opportunities
Abstract
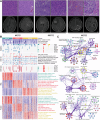
Integrating multimodal data can uncover causal features hidden in single-modality analyses, offering a comprehensive understanding of disease complexity. This study introduces a multimodal fusion subtyping (MOFS) framework that integrates radiological, pathological, genomic, transcriptomic, and proteomic data from 122 patients with IDH-wildtype adult glioma, identifying three subtypes: MOFS1 (proneural) with favorable prognosis, elevated neurodevelopmental activity, and abundant neurocyte infiltration; MOFS2 (proliferative) with the worst prognosis, superior proliferative activity, and genome instability; MOFS3 (TME-rich) with intermediate prognosis, abundant immune and stromal components, and sensitive to anti-PD-1 immunotherapy. STRAP emerges as a prognostic biomarker and potential therapeutic target for MOFS2, associated with its proliferative phenotype. Stromal infiltration in MOFS3 serves as a crucial prognostic indicator, allowing for further prognostic stratification. Additionally, we develop a deep neural network (DNN) classifier based on radiological features to further enhance the clinical translatability, providing a non-invasive tool for predicting MOFS subtypes. Overall, these findings highlight the potential of multimodal fusion in improving the classification, prognostic accuracy, and precision therapy of IDH-wildtype glioma, offering an avenue for personalized management.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

