TMEM160 inhibits KEAP1 to suppress ferroptosis and induce chemoresistance in gastric cancer
- PMID: 40223081
- PMCID: PMC11994801
- DOI: 10.1038/s41419-025-07621-0
TMEM160 inhibits KEAP1 to suppress ferroptosis and induce chemoresistance in gastric cancer
Abstract
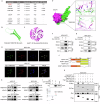
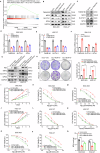
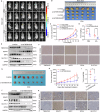
Chemoresistance is the most significant challenge affecting the clinical efficacy of the treatment of patients with gastric cancer (GC). Here we reported that transmembrane protein 160 (TMEM160) suppressed ferroptosis and induced chemoresistance in GC cells. Mechanistically, TMEM160 recruited the E3 ligase TRIM37 to promote K48-linked ubiquitination and degradation of KEAP1, thereby activating NRF2 and transcriptionally upregulating the target genes GPX4 and SLC7A11 to inhibit ferroptosis. Further in vitro and in vivo experiments demonstrated that the combination of TMEM160 targeting and chemotherapy had a synergistic inhibitory effect on the growth of GC cells, which was partially NRF2-dependent. Moreover, TMEM160 and NRF2 protein levels were markedly overexpressed in GC tissues, and their co-overexpression was an independent factor for poor prognosis. Collectively, these findings indicate that TMEM160, as a pivotal negative regulator of ferroptosis, exerts a crucial influence on the chemoresistance of GC through the TRIM37-KEAP1/NRF2 axis, providing a potential new prognostic factor and combination therapy strategy for patients with GC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: Animal experiments were approved by the animal welfare committee of the First Affiliated Hospital of Nanchang University (approval number: CDYFY-IACUC-202208QR029). All samples were approved by the Research Ethics Committee of the First Affiliated Hospital of Nanchang University (approval number: (2023)CDYFYYLK (02-068)).
Figures





References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Zheng RS, Chen R, Han BF, Wang SM, Li L, Sun KX, et al. Cancer incidence and mortality in China, 2022. Zhonghua Zhong Liu Za Zhi. 2024;46:221–31. - PubMed
-
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–48. - PubMed
-
- Noh SH, Park SR, Yang HK, Chung HC, Chung IJ, Kim SW, et al. Adjuvant capecitabine plus oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): 5-year follow-up of an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:1389–96. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

