The RBM39 degrader indisulam inhibits acute megakaryoblastic leukemia by altering the alternative splicing of ZMYND8
- PMID: 40223119
- PMCID: PMC11995665
- DOI: 10.1186/s13578-025-01380-3
The RBM39 degrader indisulam inhibits acute megakaryoblastic leukemia by altering the alternative splicing of ZMYND8
Abstract
Background: Acute megakaryoblastic leukemia (AMKL) is a rare hematological malignancy in adults but children. Alternative splicing (AS) has been shown to affect hematological cancer progression, making splicing factors promising targets. Our research aims to investigate the efficacy of the molecular glue degrader indisulam, which targets the splicing factor RNA binding motif protein 39 (RBM39) in AMKL models.
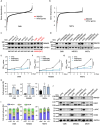
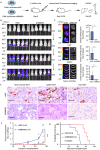
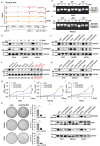
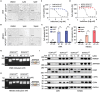
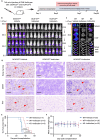
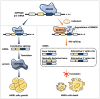
Results: Public drug sensitivity data analysis revealed that AMKL cell lines exhibited the highest sensitivity to indisulam compared with other tumor types. Then we confirmed that RBM39 depletion by indisulam treatment induced AMKL cell cycle arrest and apoptosis. In AMKL mouse model, indisulam treatment significantly reduced the leukemic burden and prolonged the lifetime of AMKL mice. Mechanically, integration of transcriptomic and proteomic analyses revealed that indisulam-mediated RBM39 degradation resulted in AS of the transcription factor zinc finger MYND-type containing 8 (ZMYND8), an AMKL cell growth regulator. Finally, the effectiveness of indisulam depended on DDB1- and Cul4- Associated Factor 15 (DCAF15) expression because knockout of DCAF15 rescued the indisulam-induced RBM39 degradation and mis-splicing of ZMYND8.
Conclusion: Indisulam is a promising therapeutic candidate for AMKL and the RBM39-mediated ZMYND8 splicing plays an important role in promoting the development of AMKL.
Keywords: Acute megakaryoblastic leukemia; Alternative splicing; Indisulam; RBM39; ZMYND8.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The animal experiments were approved by the Animal Care Committee of the Soochow University (CAM-SU-AP#: JP-2018–1), and the experiments conformed to the animal ARRIVE guidelines. Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures



Similar articles
-
Targeting RBM39 through indisulam induced mis-splicing of mRNA to exert anti-cancer effects in T-cell acute lymphoblastic leukemia.J Exp Clin Cancer Res. 2024 Jul 24;43(1):205. doi: 10.1186/s13046-024-03130-8. J Exp Clin Cancer Res. 2024. PMID: 39044280 Free PMC article.
-
Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.Nat Chem Biol. 2020 Jan;16(1):15-23. doi: 10.1038/s41589-019-0411-6. Epub 2019 Dec 9. Nat Chem Biol. 2020. PMID: 31819272
-
Recent advances in anticancer mechanisms of molecular glue degraders: focus on RBM39-dgrading synthetic sulfonamide such as indisulam, E7820, tasisulam, and chloroquinoxaline sulfonamide.Genes Genomics. 2024 Dec;46(12):1345-1361. doi: 10.1007/s13258-024-01565-z. Epub 2024 Sep 13. Genes Genomics. 2024. PMID: 39271535 Review.
-
Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15.Science. 2017 Apr 28;356(6336):eaal3755. doi: 10.1126/science.aal3755. Epub 2017 Mar 16. Science. 2017. PMID: 28302793
-
RNA-binding motif protein 39 (RBM39): An emerging cancer target.Br J Pharmacol. 2022 Jun;179(12):2795-2812. doi: 10.1111/bph.15331. Epub 2021 Jan 3. Br J Pharmacol. 2022. PMID: 33238031 Review.
References
-
- Athale UH, Razzouk BI, Raimondi SC, Tong X, Behm FG, Head DR, et al. Biology and outcome of childhood acute megakaryoblastic leukemia: a single institution’s experience. Blood. 2001;97(12):3727–32. 10.1182/blood.v97.12.3727. - PubMed
-
- Giri S, Pathak R, Prouet P, Li B, Martin MG. Acute megakaryocytic leukemia is associated with worse outcomes than other types of acute myeloid leukemia. Blood. 2014;124(25):3833–4. 10.1182/blood-2014-09-603415. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

