ATF3 Within the Interferon Signaling Pathway: A Potential Biomarker for Predicting Pathological Response to Neoadjuvant Chemoimmunotherapy
- PMID: 40223203
- PMCID: PMC11994479
- DOI: 10.1111/1759-7714.70056
ATF3 Within the Interferon Signaling Pathway: A Potential Biomarker for Predicting Pathological Response to Neoadjuvant Chemoimmunotherapy
Abstract
Background: Neoadjuvant chemoimmunotherapy has achieved high downstaging and pathologic response rates in nonsmall-cell lung cancer (NSCLC), but outcomes vary significantly. Early identification of beneficiaries remains a challenge.
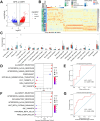
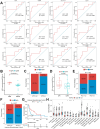
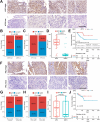
Methods: This study analyzed baseline transcriptomic data from 24 NSCLC patients (9 major pathological response [MPR], 15 nonmajor pathological response [NMPR]) treated with neoadjuvant chemoimmunotherapy, sourced from the GEO database. Molecular analyses and immune infiltration analyses were performed using pathologic response as an endpoint. After identifying the interferon signaling subset NeoIGS, we analyzed the relationship between NeoIGS and immune scores, immune cell infiltration, and immunotherapy efficacy. A key gene in NeoIGS was screened by reveiver operating characteristic curve (ROC) analysis. Subsequently, the expression of the key gene was assessed by immunohistochemistry in 53 NSCLC patients receiving neoadjuvant chemoimmunotherapy.
Results: Interferon signaling pathway expression and CD8+ T-cell infiltration were higher in the MPR group. NeoIGS predicted pathological response to neoadjuvant chemoimmunotherapy (AUC = 0.926) and also demonstrated predictive value in the ICIs monotherapy cohort. IPS and TIDE scores also confirmed NeoIGS's association with immunotherapy in the TCGA NSCLC dataset. Furthermore, patients with higher NeoIGS scores had more immune cell infiltration and increased expression of ICI targets. ROC analysis identified ATF3 as NeoIGS's key gene. In the clinical cohort, ATF3 outperformed PD-L1 in predicting pathologic response, with a 90.0% MPR rate in the high-expression group.
Conclusion: We established that a subset of interferon signaling pathways, NeoIGS, is closely associated with immunotherapy. Among them, ATF3 is the most critical gene that accurately predicts pathological remission in neoadjuvant chemoimmunotherapy.
Keywords: ATF3; IFN signaling pathway; NSCLC; biomarkers; chemoimmunotherapy; pathological response.
© 2025 The Author(s). Thoracic Cancer published by John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
A combined model using pre-treatment CT radiomics and clinicopathological features of non-small cell lung cancer to predict major pathological responses after neoadjuvant chemoimmunotherapy.Curr Probl Cancer. 2024 Jun;50:101098. doi: 10.1016/j.currproblcancer.2024.101098. Epub 2024 May 4. Curr Probl Cancer. 2024. PMID: 38704949
-
Peripheral CD8+PD-1+ T cells as novel biomarker for neoadjuvant chemoimmunotherapy in humanized mice of non-small cell lung cancer.Cancer Lett. 2024 Aug 10;597:217073. doi: 10.1016/j.canlet.2024.217073. Epub 2024 Jun 19. Cancer Lett. 2024. PMID: 38906523
-
[18F]FDG PET-CT radiomics signature to predict pathological complete response to neoadjuvant chemoimmunotherapy in non-small cell lung cancer: a multicenter study.Eur Radiol. 2024 Jul;34(7):4352-4363. doi: 10.1007/s00330-023-10503-8. Epub 2023 Dec 21. Eur Radiol. 2024. PMID: 38127071
-
A systematic review and meta-analysis of neoadjuvant chemoimmunotherapy in stage III non-small cell lung cancer.BMC Pulm Med. 2022 Dec 29;22(1):490. doi: 10.1186/s12890-022-02292-5. BMC Pulm Med. 2022. PMID: 36582007 Free PMC article.
-
Improved Event-Free Survival After Complete or Major Pathologic Response in Patients With Resectable NSCLC Treated With Neoadjuvant Chemoimmunotherapy Regardless of Adjuvant Treatment: A Systematic Review and Individual Patient Data Meta-Analysis.J Thorac Oncol. 2025 Mar;20(3):285-295. doi: 10.1016/j.jtho.2024.09.1443. Epub 2024 Oct 9. J Thorac Oncol. 2025. PMID: 39389220
References
-
- Gettinger S. N., Horn L., Gandhi L., et al., “Overall Survival and Long‐Term Safety of Nivolumab (Anti‐Programmed Death 1 Antibody, BMS‐936558, ONO‐4538) in Patients With Previously Treated Advanced Non‐Small‐Cell Lung Cancer,” Journal of Clinical Oncology 33 (2015): 2004–2012, 10.1200/JCO.2014.58.3708. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

