Intravasation-On-µDevice (INVADE): Engineering Dynamic Vascular Interfaces to Study Cancer Cell Intravasation
- PMID: 40223399
- PMCID: PMC12232236
- DOI: 10.1002/adma.202501466
Intravasation-On-µDevice (INVADE): Engineering Dynamic Vascular Interfaces to Study Cancer Cell Intravasation
Abstract
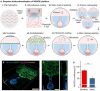
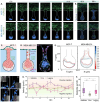
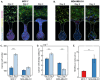
Cancer metastasis begins with intravasation, where cancer cells enter blood vessels through complex interactions with the endothelial barrier. Understanding this process remains challenging due to the lack of physiologically relevant models. Here, INVADE (Intravasation-on-µDevice), a biomimetic microfluidic platform, is presented, enabling high-throughput analysis of cancer cell intravasation under controlled conditions. This engineered platform integrates 23 parallel niche chambers with an endothelialized channel, providing both precise microenvironmental control and optical accessibility for real-time visualization. Using this platform, distinct intravasation mechanisms are uncovered: MCF-7 cells exhibit collective invasion, while MDA-MB-231 cells demonstrate an interactive mode with three functionally distinct subpopulations. A previously unknown epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET) switch is We discovered during intravasation, where MDA-MB-231 cells initially increase Vimentin expression before undergoing a 2.3 fold decrease over 96 h alongside a 1.5 fold increase in epithelial cell adhesion molecule (EpCAM). Remarkably, endothelial cells directly suppress cancer cell mesenchymal properties, as evidenced by a 4.6 fold reduction in Vimentin expression compared to mono-cultures. Additionally, bilateral cancer-endothelial interactions are revealed, aggressive cancer cells induce significant intercellular adhesion molecule-1 (ICAM-1) upregulation in endothelium. The INVADE platform represents an engineering advancement for studying complex cell-cell interactions with implications for understanding metastatic mechanisms.
Keywords: cancer metastasis; endothelium; epithelial‐mesenchymal transition; intravasation, microfluidics; vimentin.
© 2025 The Author(s). Advanced Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Chaffer C. L., Weinberg R. A., Science 2011, 331, 1559. - PubMed
-
- de Visser K. E., Joyce J. A., Cancer Cell 2023, 41, 374. - PubMed
-
- Reymond N., Agua d′ B. B., Ridley A. J., Nat. Rev. Cancer 2013, 13, 858. - PubMed
-
- Zhang Y., Jiang F., Zhao Y. C., Cho A. N., Fang G., Cox C. D., Zreiqat H., Lu Z. F., Lu H., Ju L. A., Biomed. Mater. 2023, 18, 055008. - PubMed
MeSH terms
Substances
Grants and funding
- H22/98586/NSW Cardiovascular Capacity Building Program
- DP240102315/Australian Research Council
- DP240101768/Australian Research Council
- FT230100249/Australian Research Council
- 105863/WT_/Wellcome Trust/United Kingdom
- MRF2023977/MRFF Cardiovascular Health Mission Grants
- 391-FY2023/Tour de Cure Pioneering Research
- 106979/National Heart Foundation Vanguard
- 2022SF176/Snow Medical Research Foundation
- 030GJHZ2023098FN/the International Partnership Program of Chinese Academy of Sciences
- 030GJHZ2024103MI/the International Partnership Program of Chinese Academy of Sciences
- 105863/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

