Vitamin D exerts endogenous control over TH2 cell fate and immune plasticity
- PMID: 40224021
- PMCID: PMC11987635
- DOI: 10.1016/j.isci.2025.112117
Vitamin D exerts endogenous control over TH2 cell fate and immune plasticity
Abstract
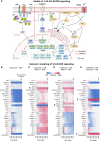
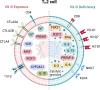
Circulating Vitamin D (Vit-D) has emerged as a potent immune modulator in asthma, yet its direct impact on TH2 cell regulation, the central effectors of allergic inflammation, remains unclear. Preliminary transcriptomic analysis of neonatal cord blood revealed that gestational Vit-D deficiency corresponds to elevated adaptive and innate immune responses, driven by TH2 immunity and antimicrobial responses related to asthma inflammation. To elucidate cell-specific molecular mechanisms of Vit-D, we differentiated murine TH2 cells in vitro under conditions mimicking Vit-D sufficiency and deficiency. Our findings demonstrate that Vit-D exposure promotes intracellular calcium ion homeostasis while suppressing prominent inflammatory cytokines characteristic of asthma. Conversely, Vit-D deficiency reprograms TH2 cell lineage commitment, inducing overexpression of cytolytic molecules and major histocompatibility complex (MHC) class I molecules-traits typically associated with cytotoxicity rather than the canonical helper function. Our findings underscore Vit-D's role in stabilizing TH2 cell function and fate, offering insights into asthma and autoimmune disorders.
Keywords: biological sciences; immune response; transcriptomics.
© 2025 The Authors.
Conflict of interest statement
Scott T. Weiss and Augusto A. Litonjua receive royalties from UpToDate. Scott T. Weiss serves as a board member of Histolix, a digital pathology company. The remaining authors declare no competing interests.
Figures





Similar articles
-
Impact of Vitamin D Supplementation on Influenza Vaccine Response and Immune Functions in Deficient Elderly Persons: A Randomized Placebo-Controlled Trial.Front Immunol. 2019 Feb 8;10:65. doi: 10.3389/fimmu.2019.00065. eCollection 2019. Front Immunol. 2019. PMID: 30800121 Free PMC article. Clinical Trial.
-
Study Effect of Vitamin D on the Immunopathology Responses of the Bronchi in Murine Model of Asthma.Iran J Allergy Asthma Immunol. 2021 Sep 28;20(5):509-519. doi: 10.18502/ijaai.v20i5.7399. Iran J Allergy Asthma Immunol. 2021. PMID: 34664810
-
The association of vitamin D, cathelicidin, and vitamin D binding protein with acute asthma attacks in children.Allergy Asthma Proc. 2015 Jul-Aug;36(4):51-8. doi: 10.2500/aap.2015.36.3848. Allergy Asthma Proc. 2015. PMID: 26108071
-
Macrophages: The Good, the Bad, and the Gluttony.Front Immunol. 2021 Aug 12;12:708186. doi: 10.3389/fimmu.2021.708186. eCollection 2021. Front Immunol. 2021. PMID: 34456917 Free PMC article. Review.
-
Understanding asthma pathogenesis: linking innate and adaptive immunity.Curr Opin Pediatr. 2004 Dec;16(6):659-66. doi: 10.1097/01.mop.0000145920.00101.e4. Curr Opin Pediatr. 2004. PMID: 15548929 Review.
Cited by
-
Gut microbiota-metabolome crosstalk in allergic diseases: mechanistic insights and translational opportunities.Front Allergy. 2025 Jul 15;6:1631479. doi: 10.3389/falgy.2025.1631479. eCollection 2025. Front Allergy. 2025. PMID: 40735411 Free PMC article. Review.
References
-
- Venkatesan P. 2023 GINA report for asthma. Lancet Respir. Med. 2023;11:589. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

