Wo interacts with SlTCP25 to regulate type I trichome branching in tomato
- PMID: 40224324
- PMCID: PMC11992337
- DOI: 10.1093/hr/uhaf032
Wo interacts with SlTCP25 to regulate type I trichome branching in tomato
Abstract
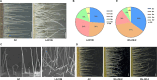
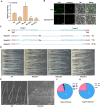
Plant trichomes serve as a protective barrier against various stresses. Although the molecular mechanisms governing the initiation of trichomes have been extensively studied, the regulatory pathways underlying the trichome branching in tomato remain elusive. Here, we found that Woolly (Wo) mutant and its overexpression transgenic plants displayed branched type I trichomes. The expression level of SlTCP25, a transcription factor of type TB1 of the TCP subfamily, was obviously decreased in Wo mutant and Wo overexpressing lines. Knockout of SlTCP25 resulted in the formation of type I trichome branches on the hypocotyls. Genetic evidence showed that SlTCP25 is epistatic to Wo in the branched trichome formation. Biochemical data further indicated that Wo can directly bind to the L1-box cis-element in the SlTCP25 promoter and repress its transcription. We further determined that SlTCP25 interacts with Wo to weaken Wo-regulated the expression of SlCycB2, a trichome branching inhibitor. In addition, the number of trichome branches was significantly increased in Sltcp25Slcycb2 double mutant, suggesting that SlTCP25 and SlCycB2 coordinately repress trichome branching in wild type. In conclusion, we elucidate a molecular network governing the morphogenesis of multicellular trichomes in tomato.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
None declared.
Figures





Similar articles
-
SlH3 and SlH4 promote multicellular trichome formation and elongation by upregulating woolly in tomato.Hortic Res. 2025 Jan 24;12(4):uhaf008. doi: 10.1093/hr/uhaf008. eCollection 2025 Apr. Hortic Res. 2025. PMID: 40093379 Free PMC article.
-
A novel regulatory complex mediated by Lanata (Ln) controls multicellular trichome formation in tomato.New Phytol. 2022 Dec;236(6):2294-2310. doi: 10.1111/nph.18492. Epub 2022 Sep 30. New Phytol. 2022. PMID: 36102042
-
Transcriptomic and functional analyses uncover the regulatory role of lncRNA000170 in tomato multicellular trichome formation.Plant J. 2020 Sep;104(1):18-29. doi: 10.1111/tpj.14902. Epub 2020 Jul 15. Plant J. 2020. PMID: 32603492
-
Molecular Mechanisms of Plant Trichome Development.Front Plant Sci. 2022 Jun 1;13:910228. doi: 10.3389/fpls.2022.910228. eCollection 2022. Front Plant Sci. 2022. PMID: 35720574 Free PMC article. Review.
-
Trichome-Related Mutants Provide a New Perspective on Multicellular Trichome Initiation and Development in Cucumber (Cucumis sativus L).Front Plant Sci. 2016 Aug 10;7:1187. doi: 10.3389/fpls.2016.01187. eCollection 2016. Front Plant Sci. 2016. PMID: 27559338 Free PMC article. Review.
References
-
- Balkunde R, Pesch M, Hülskamp M. Chapter Ten—Trichome Patterning in Arabidopsis thaliana: From Genetic to Molecular Models. In: Timmermans MCP, ed. Current Topics in Developmental Biology. Academic Press, 2010;91:299–321 - PubMed
-
- Johnson HB. Plant pubescence: An ecological perspective. Bot Rev. 1975;41:233–58
-
- Pillemer EA, Tingey WM. Hooked Trichomes: a physical plant barrier to a major agricultural Pest. Science. 1976;193:482–4 - PubMed
-
- Walker JD, Oppenheimer DG, Concienne J. et al. Trichome diversity and development. In: Advances in Botanical Research, vol. 127. Academic Press, 2000,1–35
LinkOut - more resources
Full Text Sources

