Epigenetic modification brings new opportunities for gene capture by transposable elements in allopolyploid Brassica napus
- PMID: 40224332
- PMCID: PMC11986588
- DOI: 10.1093/hr/uhaf028
Epigenetic modification brings new opportunities for gene capture by transposable elements in allopolyploid Brassica napus
Abstract
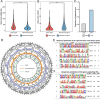
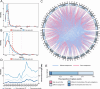
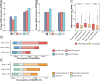
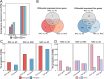
Polyploids are widespread in plants and are important drivers for evolution and biodiversity. Allopolyploidy activates transposable elements (TEs) and causes genomic shock. Plant genomes can regulate gene expression by changing the epigenetic modification of TEs, but the mechanism for TEs to capture genes remains to be explored. Helitron TEs used the 'peel-and-paste' mechanism to achieve gene capture. We identified 3156 capture events and 326 donor genes of Helitron TEs in Brassica napus (AnAnCnCn). The donor genes captured by TEs were related to the number, length, and location of their exons. The gene-capturing TEs carrying donor gene fragments were evenly distributed on the genome, and more than half of them were involved in the construction of pseudogenes, becoming the reserve force for polyploid evolution. Gene fragment copies enhanced information storage, providing opportunities for gene mutation and the formation of new genes. Simultaneously, the siRNAs targeting TEs may act on the donor genes due to siRNA crosstalk, and the gene methylation levels increased and the expression levels decreased. The genome sought a balance between sacrificing donor gene expression and silencing TEs, allowing TEs to hide in the genome. In addition, epigenetic modifications may temporarily relax the control of gene capture during allopolyploidization. Our study identified and characterized gene capture events in B. napus, analyzed the effects of DNA methylation and siRNA on gene capture events, and explored the regulation mechanism of gene expression by TE epigenetic modification during allopolyploidization, which will contribute to understanding the formation and evolution mechanism of allopolyploidy.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Genomic asymmetric epigenetic modification of transposable elements is involved in gene expression regulation of allopolyploid Brassica napus.Plant J. 2024 Jan;117(1):226-241. doi: 10.1111/tpj.16491. Epub 2023 Oct 5. Plant J. 2024. PMID: 37797206
-
Gene capture by transposable elements leads to epigenetic conflict in maize.Mol Plant. 2021 Feb 1;14(2):237-252. doi: 10.1016/j.molp.2020.11.003. Epub 2020 Nov 7. Mol Plant. 2021. PMID: 33171302
-
Impact of transposable elements on the organization and function of allopolyploid genomes.New Phytol. 2010 Apr;186(1):37-45. doi: 10.1111/j.1469-8137.2009.03096.x. Epub 2009 Dec 7. New Phytol. 2010. PMID: 20002321 Review.
-
Allopolyploidy has a moderate impact on restructuring at three contrasting transposable element insertion sites in resynthesized Brassica napus allotetraploids.New Phytol. 2013 Apr;198(2):593-604. doi: 10.1111/nph.12156. Epub 2013 Feb 6. New Phytol. 2013. PMID: 23384044
-
The Dynamism of Transposon Methylation for Plant Development and Stress Adaptation.Int J Mol Sci. 2021 Oct 21;22(21):11387. doi: 10.3390/ijms222111387. Int J Mol Sci. 2021. PMID: 34768817 Free PMC article. Review.
Cited by
-
Understanding the Regulation Activities of Transposons in Driving the Variation and Evolution of Polyploid Plant Genome.Plants (Basel). 2025 Apr 8;14(8):1160. doi: 10.3390/plants14081160. Plants (Basel). 2025. PMID: 40284048 Free PMC article. Review.
References
-
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801 - PubMed
LinkOut - more resources
Full Text Sources
Medical

