Bioinformatic Analysis of Apoptosis-Related Genes in Preeclampsia Using Public Transcriptomic and Single-Cell RNA Sequencing Datasets
- PMID: 40224388
- PMCID: PMC11992479
- DOI: 10.2147/JIR.S507660
Bioinformatic Analysis of Apoptosis-Related Genes in Preeclampsia Using Public Transcriptomic and Single-Cell RNA Sequencing Datasets
Abstract
Purpose: Apoptosis, which is crucial in preeclampsia (PE), affects trophoblast survival and placental function. We used transcriptomics and single-cell RNA sequencing (scRNA-seq) to explore apoptosis-related genes (ARGs) and their cellular mechanisms as potential PE biomarkers.
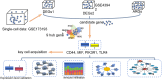
Patients and methods: All the data included in this study were sourced from public databases. We used scRNA-seq and differential expression analysis, combined with five algorithms from the CytoHubba plugin, to identify ARGs as PE biomarkers. These were integrated into diagnostic nomograms. Mechanistic studies involved enrichment analysis and immune profiling. Biomarker expression was examined at the single-cell level, and verified in clinical samples by RT-qPCR.
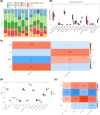
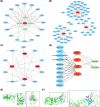
Results: Cluster of Differentiation 44 (CD44), Macrophage migration inhibitory factor (MIF), PIK3R1, and Toll-like receptor 4 (TLR4) were identified as PE biomarkers. CD44 and TLR4 were down-regulated, while MIF and PIK3R1 were up-regulated. When integrated into the diagnostic nomogram, they showed clinical utility and affected cell functions. In the immune profile of PE, monocytes decreased, resting NK cells increased, and the activities of APC, checkpoint, T-cell co-stimulation, and MHC class I pathways reduced. ScRNA-seq identified 11 cell types, 10 of which were significantly different. Endothelial cell communication with other cell types decreased, while the interaction between common myeloid progenitors (CMP) and villous cytotrophoblasts (VCT) enhanced. The expression levels of CD44, MIF, and PIK3R1 in VCT were significantly different and key to PE. Their decrease in early PE and increase in late PE reflected the placenta's adaptation to adverse pregnancy conditions.
Conclusion: Four ARGs, CD44, MIF, PIK3R1, and TLR4, identified through comprehensive analyses, served as significant biomarkers for PE and offered insights into PE's cellular mechanisms of PE, providing valuable references for further research.
Keywords: apoptosis; biomarkers; preeclampsia; single-cell RNA sequencing; villous cytotrophoblast cells.
© 2025 Liu et al.
Conflict of interest statement
The authors declare that this research was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Figures







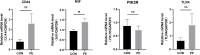
Similar articles
-
TGFβ signalling: a nexus between inflammation, placental health and preeclampsia throughout pregnancy.Hum Reprod Update. 2024 Jul 1;30(4):442-471. doi: 10.1093/humupd/dmae007. Hum Reprod Update. 2024. PMID: 38519450 Free PMC article. Review.
-
Single-cell profiling reveals immune disturbances landscape and HLA-F-mediated immune tolerance at the maternal-fetal interface in preeclampsia.Front Immunol. 2023 Oct 3;14:1234577. doi: 10.3389/fimmu.2023.1234577. eCollection 2023. Front Immunol. 2023. PMID: 37854606 Free PMC article.
-
Insights into preeclampsia: a bioinformatics approach to deciphering genetic and immune contributions.Front Genet. 2024 Aug 6;15:1372164. doi: 10.3389/fgene.2024.1372164. eCollection 2024. Front Genet. 2024. PMID: 39165753 Free PMC article.
-
Preeclampsia: novel insights from global RNA profiling of trophoblast subpopulations.Am J Obstet Gynecol. 2017 Aug;217(2):200.e1-200.e17. doi: 10.1016/j.ajog.2017.03.017. Epub 2017 Mar 24. Am J Obstet Gynecol. 2017. PMID: 28347715
-
Placenta at single-cell resolution in early and late preeclampsia: insights and clinical implications.Am J Obstet Gynecol. 2025 Apr;232(4S):S176-S189. doi: 10.1016/j.ajog.2025.01.041. Am J Obstet Gynecol. 2025. PMID: 40253080 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

