pDILI_v1: A Web-Based Machine Learning Tool for Predicting Drug-Induced Liver Injury (DILI) Integrating Chemical Space Analysis and Molecular Fingerprints
- PMID: 40224405
- PMCID: PMC11983207
- DOI: 10.1021/acsomega.5c00075
pDILI_v1: A Web-Based Machine Learning Tool for Predicting Drug-Induced Liver Injury (DILI) Integrating Chemical Space Analysis and Molecular Fingerprints
Abstract
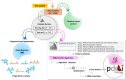
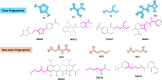
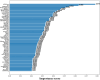
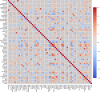
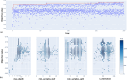
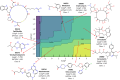
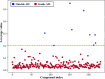
Drug-induced liver injury (DILI) represents a critical safety concern for drug development, regulatory oversight, and clinical practice, with substantial economic and public health implications. While predicting DILI risk in humans has garnered significant attention, the associated chemical space has remained insufficiently explored. This study addresses this gap through a comprehensive computational approach, leveraging machine learning (ML) to investigate structural determinants of DILI risk systematically. The study focuses on three key objectives: (i) exploring the chemical space and scaffold diversity associated with DILI; (ii) employing fragment-based approaches to identify structural alerts (SAs) that influence DILI risk; and (iii) developing supervised ML models to not only predict DILI risk but also elucidate the structural significance of molecular fingerprints. To broaden accessibility, we introduce pDILI_v1, a Python-based web application available at https://pdiliv1web.streamlit.app/. This user-friendly platform facilitates the prediction and visualization of DILI risk, enabling both experts and nonexperts to screen compounds effectively. Additional formats, including a Google Colab notebook and a graphical user interface (GUI) for Windows, ensure flexibility for diverse user needs. The proposed models demonstrate the potential for early identification of hepatotoxic risks in drug candidates, providing critical insights into drug discovery and development. By integrating ML-driven predictions with chemical space analysis, this research advances the field of drug safety evaluation, contributing to the development of safer pharmaceuticals and mitigating the risks of DILI.
© 2025 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
Prediction and mechanistic analysis of drug-induced liver injury (DILI) based on chemical structure.Biol Direct. 2021 Jan 18;16(1):6. doi: 10.1186/s13062-020-00285-0. Biol Direct. 2021. PMID: 33461600 Free PMC article.
-
Improved Detection of Drug-Induced Liver Injury by Integrating Predicted in vivo and in vitro Data.bioRxiv [Preprint]. 2024 Jun 8:2024.01.10.575128. doi: 10.1101/2024.01.10.575128. bioRxiv. 2024. Update in: Chem Res Toxicol. 2024 Aug 19;37(8):1290-1305. doi: 10.1021/acs.chemrestox.4c00015. PMID: 38895462 Free PMC article. Updated. Preprint.
-
Prediction models for drug-induced hepatotoxicity by using weighted molecular fingerprints.BMC Bioinformatics. 2017 May 31;18(Suppl 7):227. doi: 10.1186/s12859-017-1638-4. BMC Bioinformatics. 2017. PMID: 28617228 Free PMC article.
-
Computational models for predicting liver toxicity in the deep learning era.Front Toxicol. 2024 Jan 19;5:1340860. doi: 10.3389/ftox.2023.1340860. eCollection 2023. Front Toxicol. 2024. PMID: 38312894 Free PMC article. Review.
-
Control compounds for preclinical drug-induced liver injury assessment: Consensus-driven systematic review by the ProEuroDILI network.J Hepatol. 2024 Oct;81(4):630-640. doi: 10.1016/j.jhep.2024.04.026. Epub 2024 May 3. J Hepatol. 2024. PMID: 38703829
References
-
- Vaja R.; Rana M. Drugs and the liver. Anaesthesia and Intensive Care Medicine 2020, 21 (10), 517–523. 10.1016/j.mpaic.2020.07.001. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous
