Analysis and assessment of ferroptosis-related gene signatures and prognostic risk models in skin cutaneous melanoma
- PMID: 40224981
- PMCID: PMC11985187
- DOI: 10.21037/tcr-24-1506
Analysis and assessment of ferroptosis-related gene signatures and prognostic risk models in skin cutaneous melanoma
Abstract
Background: The occurrence and development of skin cutaneous melanoma (SKCM) are significantly influenced by ferroptosis, a sort of regulated cell death characterized by iron deposition and lipid peroxidation. Although positive strides have been achieved in the present management of SKCM, it is still unknown exactly how ferroptosis occurs in this condition. We aimed to determine the role of prognostically relevant ferroptosis-related genes (PR-FRGs) in SKCM development and prognosis.
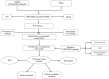
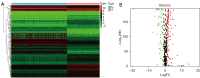
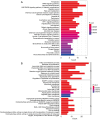
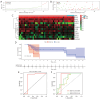
Methods: The training group was created using combined transcriptomic RNA data acquired from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases. The dataset GSE19234 was acquired from the Gene Expression Omnibus (GEO) database as a validation group. Differentially expressed ferroptosis-related genes (DE-FRGs) were obtained from the training group, of which 103 showed up-regulation and 77 showed down-regulation. Then, 12 PR-FRGs were identified by the protein-protein interaction (PPI) network and Cox regression analysis, and prognostic risk models and nomograms were constructed. The risk model was validated using a validation group, and the prognostic value of the risk model was analyzed. Finally, immunohistochemical data were obtained from the Human Protein Atlas (HPA) website to validate the PR-FRGs.
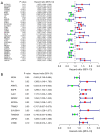
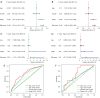
Results: Twelve PR-FRGs were identified. A prognostic risk model was built using PR-FRGs, and patients in the training and validation groups were classified as high or low risk based on the risk model. The outcomes demonstrated that the prognosis was better for the low-risk group. Prognostic value analysis showed that the prognostic risk model could accurately predict the patients' overall survival (OS), was superior to clinical traits such as age, gender, and tumor stage in predicting ability, and could be used as an independent predictor. Meanwhile, the nomogram constructed based on PR-FRGs can effectively predict the prognosis of SKCM patients. Finally, PR-FRGs were validated in the HPA database.
Conclusions: Ferroptosis affects the prognosis of SKCM patients. Prognostic risk model and nomogram constructed based on 12 PR-FRGs demonstrated significant advantages in predicting the prognosis of SKCM patients. This will help in the identification and prognostic prediction of SKCM and in the discovery of new individualized treatment modalities.
Keywords: Skin cutaneous melanoma (SKCM); ferroptosis; nomogram; prognostic model; prognostic value.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1506/coif). The authors have no conflicts of interest to declare.
Figures




Similar articles
-
Multi-omics analysis for ferroptosis-related genes as prognostic factors in cutaneous melanoma.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024 Feb 28;49(2):159-174. doi: 10.11817/j.issn.1672-7347.2024.230401. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38755712 Free PMC article.
-
A new ferroptosis-related genetic mutation risk model predicts the prognosis of skin cutaneous melanoma.Front Genet. 2023 Jan 5;13:988909. doi: 10.3389/fgene.2022.988909. eCollection 2022. Front Genet. 2023. PMID: 36685905 Free PMC article.
-
Development and validation of an immune gene set-based prognostic signature in cutaneous melanoma.Future Oncol. 2021 Nov;17(31):4115-4129. doi: 10.2217/fon-2021-0104. Epub 2021 Jul 22. Future Oncol. 2021. PMID: 34291650
-
Identification and validation of a ferroptosis-related gene signature for predicting survival in skin cutaneous melanoma.Cancer Med. 2022 Sep;11(18):3529-3541. doi: 10.1002/cam4.4706. Epub 2022 Apr 4. Cancer Med. 2022. PMID: 35373463 Free PMC article.
-
Ferroptosis-Associated Classifier and Indicator for Prognostic Prediction in Cutaneous Melanoma.J Oncol. 2021 Oct 28;2021:3658196. doi: 10.1155/2021/3658196. eCollection 2021. J Oncol. 2021. PMID: 34745259 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
