Evaluation in Monogenic Diabetes of the Impact of GCK, HNF1A, and HNF4A Variants on Splicing through the Combined Use of In Silico Tools and Minigene Assays
- PMID: 40225161
- PMCID: PMC11919142
- DOI: 10.1155/2023/6661013
Evaluation in Monogenic Diabetes of the Impact of GCK, HNF1A, and HNF4A Variants on Splicing through the Combined Use of In Silico Tools and Minigene Assays
Abstract
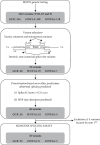
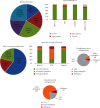
Variants in GCK, HNF1A, and HNF4A genes are the three main causes of monogenic diabetes. Determining the molecular etiology is essential for patients with monogenic diabetes to benefit from the most appropriate treatment. The increasing number of variants of unknown significance (VUS) is a major issue in genetic diagnosis, and assessing the impact of variants on RNA splicing is challenging, particularly for genes expressed in tissues not easily accessible as in monogenic diabetes. The in vitro functional splicing assay based on a minigene construct is an appropriate approach. Here, we performed in silico analysis using SpliceAI and SPiP and prioritized 36 spliceogenic variants in GCK, HNF1A, and HNF4A. Predictions were secondarily compared with Pangolin and AbSplice-DNA bioinformatics tools which include tissue-specific annotations. We assessed the effect of selected variants on RNA splicing using minigene assays. These assays validated splicing defects for 33 out of 36 spliceogenic variants consisting of exon skipping (15%), exonic deletions (18%), intronic retentions (24%), and complex splicing patterns (42%). This provided additional evidence to reclassify 23 out of 31 (74%) VUS including missense, synonymous, and intronic noncanonical splice site variants as likely pathogenic variants. Comparison of in silico analysis with minigene results showed the robustness of bioinformatics tools to prioritize spliceogenic variants, but revealed inconsistencies in the location of cryptic splice sites underlying the importance of confirming predicted splicing alterations with functional splicing assays. Our study underlines the feasibility and the benefits of implementing minigene-splicing assays in the genetic testing of monogenic diabetes after a prior in-depth in silico analysis.
Copyright © 2023 Delphine Bouvet et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


Similar articles
-
Reduced penetrance of MODY-associated HNF1A/HNF4A variants but not GCK variants in clinically unselected cohorts.Am J Hum Genet. 2022 Nov 3;109(11):2018-2028. doi: 10.1016/j.ajhg.2022.09.014. Epub 2022 Oct 17. Am J Hum Genet. 2022. PMID: 36257325 Free PMC article.
-
Mis-splicing in breast cancer: identification of pathogenic BRCA2 variants by systematic minigene assays.J Pathol. 2019 Aug;248(4):409-420. doi: 10.1002/path.5268. Epub 2019 Apr 23. J Pathol. 2019. PMID: 30883759
-
Analysis of the promoter regions of disease-causing genes in maturity-onset diabetes of the young patients.Mol Biol Rep. 2020 Sep;47(9):6759-6768. doi: 10.1007/s11033-020-05734-7. Epub 2020 Aug 28. Mol Biol Rep. 2020. PMID: 32860162
-
Tunisian Maturity-Onset Diabetes of the Young: A Short Review and a New Molecular and Clinical Investigation.Front Endocrinol (Lausanne). 2021 Jul 29;12:684018. doi: 10.3389/fendo.2021.684018. eCollection 2021. Front Endocrinol (Lausanne). 2021. PMID: 34393998 Free PMC article. Review.
-
Monogenic diabetes: a gateway to precision medicine in diabetes.J Clin Invest. 2021 Feb 1;131(3):e142244. doi: 10.1172/JCI142244. J Clin Invest. 2021. PMID: 33529164 Free PMC article. Review.
References
-
- Saint-Martin C., Bouvet D., Bastide M., Chantelot C. B. Monogenic Diabetes Study Group of the Société Francophone du Diabète. Gene panel sequencing of patients with monogenic diabetes brings to light genes typically associated with syndromic presentations. Diabetes . 2022;71(3):578–584. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

