Phenotypic Diversity in GNAO1 Patients: A Comprehensive Overview of Variants and Phenotypes
- PMID: 40225165
- PMCID: PMC11919132
- DOI: 10.1155/2023/6628283
Phenotypic Diversity in GNAO1 Patients: A Comprehensive Overview of Variants and Phenotypes
Abstract
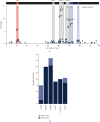
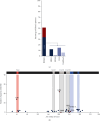
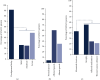
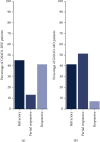
GNAO1 disorder is a rare autosomal dominant neurodevelopmental syndrome that is clinically manifested by developmental delay, (early onset) epilepsy, and movement disorders. Clinical symptoms appear very heterogeneous in nature and severity, as well as the response of GNAO1 patients to available medication varies. Pathogenic GNAO1 variants have been found mainly scattered throughout the gene although certain mutation hotspots affecting the function of the encoded Gαo proteins exist. GNAO1 variants only partially explain the diverse phenotypic spectrum observed but full stratification has been hampered by the limited number of patients. The aim of this review was to generate a comprehensive overview of the germline variants in GNAO1 and provide insight into the phenotypic diversity of the GNAO1 disorder. We compiled a list of 398 GNAO1 germline variants. In addition, we provide the GNAO1 variants and associated phenotypes of 282 GNAO1 patients reported in case reports, whole genome sequencing studies, genetic variant databases, and 8 novel GNAO1 patients that were not described before. This has resulted in a list of 107 (likely) pathogenic GNAO1 variants. Available phenotypic data was utilized to quantitatively assess the genetic and phenotypic diversity of the GNAO1 disorder and discuss the outcomes. This inventory forms the basis for a GNAO1 variant database that will be updated continuously. Moreover, it will aid genetic diagnostics, medical decision-making, prognostication, and research on the mechanisms underlying the GNAO1 disorder.
Copyright © 2023 Maria Sáez González et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




References
-
- Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine . 2015;17(5):405–424. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
-
- Offermanns S., Rosenthal W. Encyclopedia of Molecular Pharmacology . Berlin: Springer; 2008. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

