The Broad Spectrum of TP53 Mutations in CLL: Evidence of Multiclonality and Novel Mutation Hotspots
- PMID: 40225169
- PMCID: PMC11918887
- DOI: 10.1155/2023/4880113
The Broad Spectrum of TP53 Mutations in CLL: Evidence of Multiclonality and Novel Mutation Hotspots
Abstract
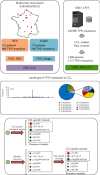
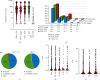
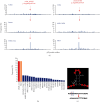
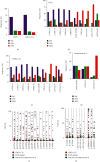
TP53 aberrations are a major predictive factor of resistance to chemoimmunotherapy in chronic lymphocytic leukemia (CLL), and an assessment of them before each line of treatment is required for theranostic stratification. Acquisition of subclonal TP53 abnormalities underlies the evolution of CLL. To better characterize the distribution, combination, and impact of TP53 variants in CLL, 1,056 TP53 variants collected from 683 patients included in a multicenter collaborative study in France were analyzed and compared to UMD_CLL, a dataset built from published articles collectively providing 5,173 TP53 variants detected in 3,808 patients. Our analysis confirmed the presence of several CLL-specific hotspot mutations, including a two-base pair deletion in codon 209 and a missense variant at codon 234, the latter being associated with alkylating treatment. Our analysis also identified a novel CLL-specific variant in the splice acceptor signal of intron 6 leading to the use of a cryptic splice site, similarly utilized by TP53 to generate p53psi, a naturally truncated p53 isoform localized in the mitochondria. Examination of both UMD_CLL and several recently released large-scale genomic analyses of CLL patients confirmed that this splice variant is highly enriched in this disease when compared to other cancer types. Using a TP53-specific single-nucleotide polymorphism, we also confirmed that copy-neutral loss of heterozygosity is frequent in CLL. This event can lead to misinterpretation of TP53 status. Unlike other cancers, CLL displayed a high proportion of patients harboring multiple TP53 variants. Using both in silico analysis and single molecule smart sequencing, we demonstrated the coexistence of distinct subclones harboring mutations on distinct alleles. In summary, our study provides a detailed TP53 mutational architecture in CLL and gives insights into how treatments may shape the genetic landscape of CLL patients.
Copyright © 2023 Grégory Lazarian et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




References
-
- Gaidano G., Ballerini P., Gong J. Z., et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America . 1991;88(12):5413–5417. doi: 10.1073/pnas.88.12.5413. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

