Colorectal cancer stem cells develop NK cell resistance via homotypic cell-in-cell structures suppressed by Stathmin1
- PMID: 40225568
- PMCID: PMC11984389
- DOI: 10.7150/thno.110379
Colorectal cancer stem cells develop NK cell resistance via homotypic cell-in-cell structures suppressed by Stathmin1
Abstract
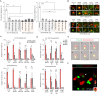
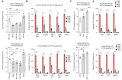
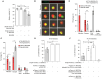
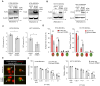
Rationale: Advances in cancer therapies have significantly improved patient survival; however, tumors enriched in cancer stem cells (CSCs) have poor treatment responses. CSCs are a key source of tumor heterogeneity, contributing to therapeutic resistance and unfavorable patient outcomes. In the tumor microenvironment (TME), cell-in-cell (CIC) structures, where one cell engulfs another, have been identified as markers of poor prognosis. Despite their clinical relevance, the mechanisms underlying CIC formation across different tumor cell subpopulations remain largely unknown. Elucidating these processes could provide novel insights and therapeutic opportunities to address aggressive, treatment-resistant cancers. Method: Fluorescent mCherry-carrying colorectal cancer stem cells (CRCSCs) were expanded as spheroids in serum-free media and cocultured with either parental cancer cell-expressing Venus fluorescent protein or CFSE dye-stained immune cells (T cells, M1/M2 macrophages, neutrophils, and NK cells) or treated with EGFR- or PD-L1-targeting antibodies to assess the formation of CIC structures. Genes potentially crucial for the formation of CIC structures were knocked down or overexpressed, and their effects on CIC formation were evaluated. The clinical relevance of the in vitro findings was confirmed through analysis of formalin-fixed, paraffin-embedded (FFPE) human colorectal cancer (CRC) specimens. Results: CRCSCs have a strong predilection for serving as the outer cell in a CIC structure and forming homotypic CIC structures predominantly with parental CRC cells. The frequency of CIC structure formation increased when the cells were exposed to anti-PD-L1 antibody treatment. Both the outer CRCSC in a CIC structure and CRCSCs released from a homotypic CIC structure showed enhanced resistance to the cytotoxicity of NK-92MI cells. Restoration of Stathmin1 (STMN1) expression but not RAC1 knockdown in CRCSCs reduced the homotypic CIC frequency, disrupted the outer cell fate in CIC structures, and increased cell susceptibility to NK-92MI cytotoxicity. In CRC patients, CIC structures are associated with poor tumor differentiation, negative STMN1 expression, and poor prognosis. Conclusion: CSCs play a crucial role in informing CIC structures in CRC. CIC structure formation partially depends on low STMN1 expression and confers a survival advantage under NK cytotoxicity. Targeting this pathway may significantly improve immunotherapy's efficacy for CRC patients.
Keywords: Cell-in-Cell Structure; Colorectal Cancer Stem Cell; Immunotherapy; Stathmin1; Tumor Microenvironment.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures

References
-
- Overholtzer M, Brugge JS. The cell biology of cell-in-cell structures. Nat Rev Mol Cell Biol. 2008;9:796–809. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

