FOS-driven inflammatory CAFs promote colorectal cancer liver metastasis via the SFRP1-FGFR2-HIF1 axis
- PMID: 40225580
- PMCID: PMC11984394
- DOI: 10.7150/thno.111625
FOS-driven inflammatory CAFs promote colorectal cancer liver metastasis via the SFRP1-FGFR2-HIF1 axis
Abstract
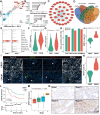
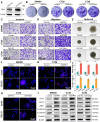
Rationale: Cancer-associated fibroblasts (CAFs) exhibit diverse functions, yet their roles in colorectal cancer liver metastasis (CRLM) remain poorly understood. Methods: Through integrated analysis of single-cell RNA sequencing and spatial transcriptomics from colorectal cancer patients (CRCP: non-metastatic primary tumors; CRCM: metastatic primary tumors with liver metastases), combined with in vitro and in vivo models to investigate the role of CAFs in CRLM. In vitro experiments included six groups to reveal the role of SFRP1-producing CAFs, comprising PBS (control) and recombinant human SFRP1 (rhSFRP1) treated SW480 cells, PBS (control) and recombinant mouse SFRP1 (rmSFRP1) treated CT26 cells, and conditioned medium (CM) derived from CAF-NC and CAF-Sfrp1 treated CT26 cells. Preclinical models were further employed to elucidate the role of SFRP1 in CRLM. Subcutaneous xenografts models were constructed from PBS (control) and rhSFRP1 treated SW480 cells. For orthotopic tumor metastasis models, CT26 cells were pre-cultured with CAF-NC or CAF-Sfrp1 and then orthotopically injected into BALB/c mice. Results: We identified an inflammatory CAF subtype (CFD+ iCAFs) associated with poor clinical outcomes, advanced staging, and metastasis. Transcriptional regulation analysis revealed FOS-mediated differentiation of CFD+ iCAFs drives SFRP1 overexpression. In vitro and in vivo experiments confirmed that SFRP1-producing CAFs promote tumor stemness and epithelial-mesenchymal transition (EMT). Mechanistically, SFRP1 from CFD+ iCAFs binds FGFR2, activating the HIF1 signaling pathway to enhance tumor stemness, EMT, and CRLM progression. Conclusion: This study highlights CFD+ iCAFs as key regulators of tumor-stromal interactions and identifies SFRP1 as a potential therapeutic target in CRLM.
Keywords: Cancer-associated fibroblasts; Colorectal cancer liver metastasis; SFRP1; Single-cell RNA sequencing; Therapeutic target.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Canellas-Socias A, Sancho E, Batlle E. Mechanisms of metastatic colorectal cancer. Nat Rev Gastroenterol Hepatol. 2024;21:609–25. - PubMed
-
- Cords L, de Souza N, Bodenmiller B. Classifying cancer-associated fibroblasts-The good, the bad, and the target. Cancer Cell. 2024;42:1480–5. - PubMed
-
- Liu Y, Zhang X, Gu W, Su H, Wang X, Wang X. et al. Unlocking the crucial role of cancer-associated fibroblasts in tumor metastasis: Mechanisms and therapeutic prospects. J Adv Res. 2024;31:S2090–1232. (24)00220-0. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

