Treatability of the KMT2-Associated Neurodevelopmental Disorders Using Antisense Oligonucleotide-Based Treatments
- PMID: 40225946
- PMCID: PMC11925151
- DOI: 10.1155/2024/9933129
Treatability of the KMT2-Associated Neurodevelopmental Disorders Using Antisense Oligonucleotide-Based Treatments
Abstract
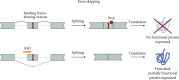
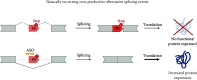
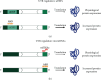
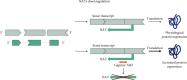
Neurodevelopmental disorders (NDDs) of genetic origin are a group of early-onset neurological diseases with highly heterogeneous etiology and a symptomatic spectrum that includes intellectual disability, autism spectrum disorder, and learning and language disorders. One group of rare NDDs is associated with dysregulation of the KMT2 protein family. Members of this family share a common methyl transferase function and are involved in the etiology of rare haploinsufficiency disorders. For each of the KMT2 genes, at least one distinct disorder has been reported, yet clinical manifestations often overlap for multiple of these individually very rare disorders. Clinical care is currently focused on the management of symptoms with no targeted treatments available, illustrating a high unmet medical need and the urgency of developing disease-modifying therapeutic strategies. Antisense oligonucleotides (ASOs) are one option to treat some of these rare genetic disorders. ASOs are RNA-based treatments that can be employed to modulate gene expression through various mechanisms. In this work, we discuss the phenotypic features across the KMT2-associated NDDs and which ASO approaches are most suited for the treatment of each associated disorder. We hereby address variant-specific strategies as well as options applicable to larger groups of patients.
Copyright © 2024 Bianca Zardetto et al.
Conflict of interest statement
AAR discloses being employed by LUMC which has patents on exon skipping technology, some of which has been licensed to BioMarin and subsequently sublicensed to Sarepta. As coinventor of some of these patents, AAR is entitled to a share of royalties. AAR further discloses being ad hoc consultant for PTC Therapeutics, Sarepta Therapeutics, Regenxbio, Alpha Anomeric, Lilly BioMarin Pharmaceuticals Inc., Eisai, Entrada, Takeda, Splicesense, Galapagos, and Astra Zeneca. Past ad hoc consulting has occurred for CRISPR Therapeutics, Summit PLC, Audentes Santhera, Bridge Bio, Global Guidepoint and GLG consultancy, Grunenthal, Wave, and BioClinica. AAR also reports having been a member of the Duchenne Network Steering Committee (BioMarin) and being a member of the scientific advisory boards of Eisai, hybridize therapeutics, silence therapeutics, Sarepta therapeutics. AAR has past SAB memberships of the following: ProQR and Philae Pharmaceuticals. Remuneration for these activities is paid to LUMC. LUMC also received speaker honoraria from PTC Therapeutics, Alnylam Netherlands, Pfizer, and BioMarin Pharmaceuticals and funding for contract research from Italfarmaco, Sapreme, Eisai, Galapagos, Synaffix, and Alpha Anomeric. Project funding is received from Sarepta Therapeutics and Entrada. WvRM discloses being employed by LUMC which has patents on exon skipping approaches for neurological disorders. In the past, some of these patents have been licensed to ProQR therapeutics. As coinventor on these patents, WvRM is entitled to a share of milestone payments and royalties. WvRM further discloses being ad hoc consultant for Accure Therapeutics and Herbert Smith Freehills. Remuneration for these activities is paid to the LUMC. LUMC also received funding for contract research from UniQure and Amylon Therapeutics.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

