Maize leaves salt-responsive genes revealed by comparative transcriptome of salt-tolerant and salt-sensitive cultivars during the seedling stage
- PMID: 40226543
- PMCID: PMC11994069
- DOI: 10.7717/peerj.19268
Maize leaves salt-responsive genes revealed by comparative transcriptome of salt-tolerant and salt-sensitive cultivars during the seedling stage
Abstract
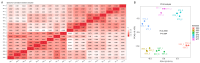
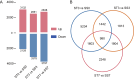
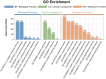
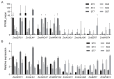
Maize (Zea mays) is a crop of significant global importance, yet its productivity is considerably hindered by salt stress. In this study, we investigated two maize cultivars, one exhibiting high salt tolerance (ST) and the other showing salt sensitivity (SS) at the seedling stage. The ST cultivar demonstrated superior seedling survival rates, higher relative water content, and lower electrolyte leakage and malondialdehyde levels in its leaves after both 3-day and 7-day salt treatments, when compared to the SS cultivar. To explore the molecular basis of these differences, we performed comparative transcriptome sequencing under varying salt treatment durations. A total of 980 differentially expressed genes (DEGs) were identified. Gene ontology (GO) functional enrichment analysis of DEGs indicated that the oxidation-reduction process, phosphorylation, plasma membrane, transferase activity, metal ion binding, kinase activity, protein kinase activity and oxidoreductase activity process is deeply involved in the response of ST and SS maize varieties to salt stress. Further analysis highlighted differences in the regulatory patterns of transcription factors encoded by the DEGs between the ST and SS cultivars. Notably, transcription factor families such as AP2/ERF, bZIP, MYB, and WRKY were found to play crucial roles in the salt stress regulatory network of maize. These findings provide valuable insights into the molecular mechanisms underlying salt stress tolerance in maize seedlings.
Keywords: Differentially expressed genes; Maize seedling leaves; RNA sequencing; Salt stress; Salt tolerance.
©2025 Ji et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



References
-
- Ding A, Tang X, Yang D, Wang M, Ren A, Xu Z, Hu R, Zhou G, O’Neill M, Kong Y. ERF4 and MYB52 transcription factors play antagonistic roles in regulating homogalacturonan de-methylesterification in Arabidopsis seed coat mucilage. The Plant Cell. 2021;33:381–403. doi: 10.1093/plcell/koaa031. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

