Using Nano-Luciferase Binary (NanoBiT) Technology to Assess the Interaction Between Viral Spike Protein and Angiotensin-Converting Enzyme II by Aptamers
- PMID: 40227272
- PMCID: PMC11940275
- DOI: 10.3390/biotech14010020
Using Nano-Luciferase Binary (NanoBiT) Technology to Assess the Interaction Between Viral Spike Protein and Angiotensin-Converting Enzyme II by Aptamers
Abstract
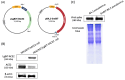
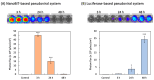
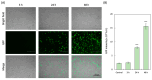
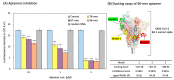
Nano-luciferase binary technology (NanoBiT)-based pseudoviral sensors are innovative tools for monitoring viral infection dynamics. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infects host cells via its trimeric surface spike protein, which binds to the human angiotensin-converting enzyme II (hACE2) receptor. This interaction is crucial for viral entry and serves as a key target for therapeutic interventions against coronavirus disease 2019 (COVID-19). Aptamers, short single-stranded DNA (ssDNA) or RNA molecules, are highly specific, high-affinity biorecognition elements for detecting infective pathogens. Despite their potential, optimizing viral infection assays using traditional protein-protein interaction (PPI) methods often face challenges in optimizing viral infection assays. In this study, we selected and evaluated aptamers for their ability to interact with viral proteins, enabling the dynamic visualization of infection progression. The NanoBiT-based pseudoviral sensor demonstrated a rapid increase in luminescence within 3 h, offering a real-time measure of viral infection. A comparison of detection technologies, including green fluorescent protein (GFP), luciferase, and NanoBiT technologies for detecting PPI between the pseudoviral spike protein and hACE2, highlighted NanoBiT's superior sensitivity and performance, particularly in aptamer selection. This bioluminescent system provides a robust, sensitive, and early-stage quantitative approach to studying viral infection dynamics.
Keywords: angiotensin-converting enzyme type II (ACE2); aptamer; nano-luciferase binary technology (NanoBiT); severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); spike protein.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Almulla N., Soltane R., Alasiri A., Allayeh A.K., Alqadi T., Alshehri F., Hamad Alrokban A., Zaghlool S.S., Zayan A.Z., Abdalla K.F., et al. Advancements in SARS-CoV-2 detection: Navigating the molecular landscape and diagnostic technologies. Heliyon. 2024;10:e29909. doi: 10.1016/j.heliyon.2024.e29909. - DOI - PMC - PubMed
-
- Dixon A.S., Schwinn M.K., Hall M.P., Zimmerman K., Otto P., Lubben T.H., Butler B.L., Binkowski B.F., Machleidt T., Kirkland T.A., et al. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem. Biol. 2016;11:400–408. doi: 10.1021/acschembio.5b00753. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
