Structure-Based Modeling of the Gut Bacteria-Host Interactome Through Statistical Analysis of Domain-Domain Associations Using Machine Learning
- PMID: 40227324
- PMCID: PMC11940256
- DOI: 10.3390/biotech14010013
Structure-Based Modeling of the Gut Bacteria-Host Interactome Through Statistical Analysis of Domain-Domain Associations Using Machine Learning
Abstract
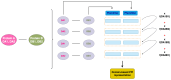
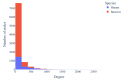
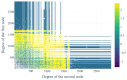
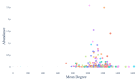
The gut microbiome, a complex ecosystem of microorganisms, plays a pivotal role in human health and disease. The gut microbiome's influence extends beyond the digestive system to various organs, and its imbalance is linked to a wide range of diseases, including cancer and neurodevelopmental, inflammatory, metabolic, cardiovascular, autoimmune, and psychiatric diseases. Despite its significance, the interactions between gut bacteria and human proteins remain understudied, with less than 20,000 experimentally validated protein interactions between the host and any bacteria species. This study addresses this knowledge gap by predicting a protein-protein interaction network between gut bacterial and human proteins. Using statistical associations between Pfam domains, a comprehensive dataset of over one million experimentally validated pan-bacterial-human protein interactions, as well as inter- and intra-species protein interactions from various organisms, were used for the development of a machine learning-based prediction method to uncover key regulatory molecules in this dynamic system. This study's findings contribute to the understanding of the intricate gut microbiome-host relationship and pave the way for future experimental validation and therapeutic strategies targeting the gut microbiome interplay.
Keywords: domain interactions; gut microbiome; host–bacteria interactions; machine learning; protein networks.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous
