Chromosomal Instability and Clonal Heterogeneity in Breast Cancer: From Mechanisms to Clinical Applications
- PMID: 40227811
- PMCID: PMC11988187
- DOI: 10.3390/cancers17071222
Chromosomal Instability and Clonal Heterogeneity in Breast Cancer: From Mechanisms to Clinical Applications
Abstract
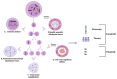
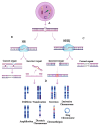
Background: Chromosomal instability (CIN) and clonal heterogeneity (CH) are fundamental hallmarks of breast cancer that drive tumor evolution, disease progression, and therapeutic resistance. Understanding the mechanisms underlying these phenomena is essential for improving cancer diagnosis, prognosis, and treatment strategies.
Methods: In this review, we provide a comprehensive overview of the biological processes contributing to CIN and CH, highlighting their molecular determinants and clinical relevance.
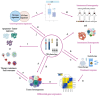
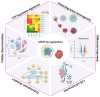
Results: We discuss the latest advances in detection methods, including single-cell sequencing and other high-resolution techniques, which have enhanced our ability to characterize intratumoral heterogeneity. Additionally, we explore how CIN and CH influence treatment responses, their potential as therapeutic targets, and their role in shaping the tumor immune microenvironment, which has implications for immunotherapy effectiveness.
Conclusions: By integrating recent findings, this review underscores the impact of CIN and CH on breast cancer progression and their translational implications for precision medicine.
Keywords: aneuploidy; breast cancer; chromosomal instability; clonal heterogeneity; gene expression; single-cell sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Patterns of Chromosomal Instability and Clonal Heterogeneity in Luminal B Breast Cancer: A Pilot Study.Int J Mol Sci. 2024 Apr 19;25(8):4478. doi: 10.3390/ijms25084478. Int J Mol Sci. 2024. PMID: 38674062 Free PMC article.
-
Exploring chromosomal instability and clonal heterogeneity in breast cancer.Endocr Relat Cancer. 2024 Nov 5;31(12):e240096. doi: 10.1530/ERC-24-0096. Print 2024 Dec 1. Endocr Relat Cancer. 2024. PMID: 39315955
-
Chromosomal Instability as Enabling Feature and Central Hallmark of Breast Cancer.Breast Cancer (Dove Med Press). 2023 Mar 9;15:189-211. doi: 10.2147/BCTT.S383759. eCollection 2023. Breast Cancer (Dove Med Press). 2023. PMID: 36923397 Free PMC article. Review.
-
Chromosome Instability; Implications in Cancer Development, Progression, and Clinical Outcomes.Cancers (Basel). 2020 Mar 29;12(4):824. doi: 10.3390/cancers12040824. Cancers (Basel). 2020. PMID: 32235397 Free PMC article. Review.
-
Role of chromosomal instability and clonal heterogeneity in the therapy response of breast cancer cell lines.Cancer Biol Med. 2020 Nov 15;17(4):970-985. doi: 10.20892/j.issn.2095-3941.2020.0028. Epub 2020 Dec 15. Cancer Biol Med. 2020. PMID: 33299647 Free PMC article.
References
-
- Hanahan D., Weinberg R.A. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. - PubMed
-
- Lengauer C., Kinzler K.W., Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. - PubMed
-
- Sansregret L., Vanhaesebroeck B., Swanton C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018;15:139–150. - PubMed
-
- McGranahan N., Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 2017;168:613–628. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

