FDG-PET/CT and Multimodal Machine Learning Model Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer
- PMID: 40227836
- PMCID: PMC11987901
- DOI: 10.3390/cancers17071249
FDG-PET/CT and Multimodal Machine Learning Model Prediction of Pathological Complete Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer
Abstract
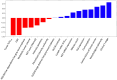
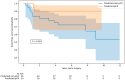
Purpose: Triple-negative breast cancer (TNBC) is a biologically and clinically heterogeneous disease, associated with poorer outcomes when compared with other subtypes of breast cancer. Neoadjuvant chemotherapy (NAC) is often given before surgery, and achieving a pathological complete response (pCR) has been associated with patient outcomes. There is thus strong clinical interest in the ability to accurately predict pCR status using baseline data. Materials and Methods: A cohort of 57 TNBC patients who underwent FDG-PET/CT before NAC was analyzed to develop a machine learning (ML) algorithm predictive of pCR. A total of 241 predictors were collected for each patient: 11 clinical features, 11 histopathological features, 13 genomic features, and 206 PET features, including 195 radiomic features. The optimization criterion was the area under the ROC curve (AUC). Event-free survival (EFS) was estimated using the Kaplan-Meier method. Results: The best ML algorithm reached an AUC of 0.82. The features with the highest weight in the algorithm were a mix of PET (including radiomics), histopathological, genomic, and clinical features, highlighting the importance of truly multimodal analysis. Patients with predicted pCR tended to have a longer EFS than patients with predicted non-pCR, even though this difference was not significant, probably due to the small sample size and few events observed (p = 0.09). Conclusions: This study suggests that ML applied to baseline multimodal data can help predict pCR status after NAC for TNBC patients and may identify correlations with long-term outcomes. Patients predicted as non-pCR may benefit from concomitant treatment with immunotherapy or dose intensification.
Keywords: FDG-PET/CT; artificial intelligence; machine learning; metabolic response; neoadjuvant chemotherapy; pCR; prognosis; radiomics; triple-negative breast cancer.
Conflict of interest statement
Authors Loïc Ferrer, Jennifer Vargas, Philippe Menu, Olivier Gallinato and Thierry Colin were employed by the company SOPHiA GENETICS. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- NCCN Clinical Practice Guidelines in Oncology. Breast Cancer. Version 3. 2025. [(accessed on 3 April 2025)]. Available online: https://www.NCCN.Org/Professionals/Physician_gls/Pdf/Breast.Pdf.
-
- Gralow J.R., Burstein H.J., Wood W., Hortobagyi G.N., Gianni L., von Minckwitz G., Buzdar A.U., Smith I.E., Symmans W.F., Singh B., et al. Preoperative Therapy in Invasive Breast Cancer: Pathologic Assessment and Systemic Therapy Issues in Operable Disease. J. Clin. Oncol. 2008;26:814–819. doi: 10.1200/JCO.2007.15.3510. - DOI - PubMed
-
- Cortazar P., Zhang L., Untch M., Mehta K., Costantino J.P., Wolmark N., Bonnefoi H., Cameron D., Gianni L., Valagussa P., et al. Pathological Complete Response and Long-Term Clinical Benefit in Breast Cancer: The CTNeoBC Pooled Analysis. Lancet. 2014;384:164–172. doi: 10.1016/S0140-6736(13)62422-8. - DOI - PubMed
-
- Coudert B., Pierga J.-Y., Mouret-Reynier M.-A., Kerrou K., Ferrero J.-M., Petit T., Kerbrat P., Dupré P.-F., Bachelot T., Gabelle P., et al. Use of [(18)F]-FDG PET to Predict Response to Neoadjuvant Trastuzumab and Docetaxel in Patients with HER2-Positive Breast Cancer, and Addition of Bevacizumab to Neoadjuvant Trastuzumab and Docetaxel in [(18)F]-FDG PET-Predicted Non-Responders (AVATAXHER): An Open-Label, Randomised Phase 2 Trial. Lancet Oncol. 2014;15:1493–1502. doi: 10.1016/S1470-2045(14)70475-9. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

