Limitations and optimizations of cellular lineages tracking
- PMID: 40228207
- PMCID: PMC11996212
- DOI: 10.1371/journal.pcbi.1012880
Limitations and optimizations of cellular lineages tracking
Abstract
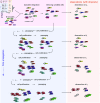
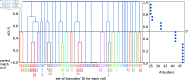
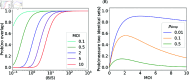
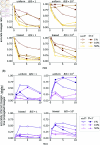
Tracking cellular lineages using genetic barcodes provides insights across biology and has become an important tool. However, barcoding strategies remain ad hoc. We show that elevating barcode insertion probability and thus increasing the average number of barcodes within the cells, adds to the number of traceable lineages but may decrease the accuracy of lineages inference due to reading errors. We establish the trade-off between accuracy in tracing lineages and the total number of traceable lineages, and find optimal experimental parameters under limited resources concerning the populations size of tracked cells and barcode pool complexity.
Copyright: © 2025 Leibovich and Goyal. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Cellular barcoding: lineage tracing, screening and beyond.Nat Methods. 2018 Nov;15(11):871-879. doi: 10.1038/s41592-018-0185-x. Epub 2018 Oct 30. Nat Methods. 2018. PMID: 30377352 Review.
-
Single-Cell Lineage Tracing and Clonal State-Fate Analysis.Methods Mol Biol. 2025;2886:65-84. doi: 10.1007/978-1-0716-4310-5_4. Methods Mol Biol. 2025. PMID: 39745636
-
Machine-learning-optimized Cas12a barcoding enables the recovery of single-cell lineages and transcriptional profiles.Mol Cell. 2022 Aug 18;82(16):3103-3118.e8. doi: 10.1016/j.molcel.2022.06.001. Epub 2022 Jun 24. Mol Cell. 2022. PMID: 35752172 Free PMC article.
-
Clonal tracking using embedded viral barcoding and high-throughput sequencing.Nat Protoc. 2020 Apr;15(4):1436-1458. doi: 10.1038/s41596-019-0290-z. Epub 2020 Mar 4. Nat Protoc. 2020. PMID: 32132718 Free PMC article.
-
Advancements in prospective single-cell lineage barcoding and their applications in research.Genome Res. 2024 Dec 23;34(12):2147-2162. doi: 10.1101/gr.278944.124. Genome Res. 2024. PMID: 39572229 Free PMC article. Review.
Cited by
-
Barcodes based on nucleic acid sequences: Applications and challenges (Review).Mol Med Rep. 2025 Jul;32(1):187. doi: 10.3892/mmr.2025.13552. Epub 2025 May 2. Mol Med Rep. 2025. PMID: 40314098 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

