Viral pseudo-enzyme facilitates KSHV lytic replication via suppressing PFAS-mediated RTA deamidation
- PMID: 40228741
- PMCID: PMC12282450
- DOI: 10.1016/j.virs.2025.04.005
Viral pseudo-enzyme facilitates KSHV lytic replication via suppressing PFAS-mediated RTA deamidation
Abstract
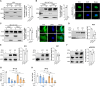
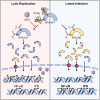
Deamidation, a type of post-translational modification commonly considered a hallmark of protein "aging" and function decay, is increasingly recognized for its pivotal role in regulating biological processes and viral infection. Our previous study has demonstrated that the deamidation of replication and transcription activator (RTA), a master regulator of ubiquitous and oncogenic Kaposi's sarcoma-associated herpesvirus (KSHV), mediated by phosphoribosylformylglycinamidine synthetase (PFAS), hinders its nuclear import and transcriptional activity. Here we report that the viral glutamine amidotransferase (vGAT) pseudo-enzyme is exploited to facilitate KSHV lytic infection by inhibiting RTA deamidation. To be more specific, vGAT interacts with both RTA and cellular PFAS, and inhibits PFAS-mediated RTA deamidation, thus facilitating RTA nuclear localization and suppressing nuclear factor-kappa B (NF-κB) signaling activation, as well as augmenting RTA-mediated transcriptional activation of viral open reading frames (ORFs). In addition, vGAT appears to regulate the deamidation process of several viral ORFs of KSHV. Collectively, these findings unveil that a viral pseudo-enzyme is exploited to enhance viral infection via deamidation regulation.
Keywords: Deamidation; Kaposi's sarcoma-associated herpesvirus (KSHV); transcription activator (RTA); viral glutamine amidotransferase (vGAT).
Copyright © 2025 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest.
Figures





References
-
- Arias C., Weisburd B., Stern-Ginossar N., Mercier A., Madrid A.S., Bellare P., Holdorf M., Weissman J.S., Ganem D. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014;10 - PMC - PubMed
-
- Chang L.K., Lee Y.H., Cheng T.S., Hong Y.R., Lu P.J., Wang J.J., Wang W.H., Kuo C.W., Li S.S., Liu S.T. Post-translational modification of Rta of Epstein-Barr virus by SUMO-1. J. Biol. Chem. 2004;279:38803–38812. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

